Single-chromosome Gains Commonly Function as Tumor Suppressors
- PMID: 28089890
- PMCID: PMC5713901
- DOI: 10.1016/j.ccell.2016.12.004
Single-chromosome Gains Commonly Function as Tumor Suppressors
Abstract
Aneuploidy is a hallmark of cancer, although its effects on tumorigenesis are unclear. Here, we investigated the relationship between aneuploidy and cancer development using cells engineered to harbor single extra chromosomes. We found that nearly all trisomic cell lines grew poorly in vitro and as xenografts, relative to genetically matched euploid cells. Moreover, the activation of several oncogenic pathways failed to alleviate the fitness defect induced by aneuploidy. However, following prolonged growth, trisomic cells acquired additional chromosomal alterations that were largely absent from their euploid counterparts and that correlated with improved fitness. Thus, while single-chromosome gains can suppress transformation, the genome-destabilizing effects of aneuploidy confer an evolutionary flexibility that may contribute to the aggressive growth of advanced malignancies with complex karyotypes.
Keywords: aneuploidy; chromosomal instability; genome dosage imbalance; genomic instability; transformation.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures



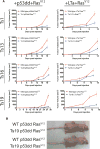

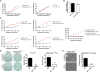

Comment in
-
Tumorigenesis: Fitness penalties of aneuploidy.Nat Rev Cancer. 2017 Mar;17(3):142-143. doi: 10.1038/nrc.2017.9. Epub 2017 Feb 10. Nat Rev Cancer. 2017. PMID: 28184042 No abstract available.
-
Singling Out Chromosome Gains in Tumor Evolution.Cancer Cell. 2017 Feb 13;31(2):165-166. doi: 10.1016/j.ccell.2017.01.011. Cancer Cell. 2017. PMID: 28196590 Free PMC article.
References
-
- Ahuja D, Sáenz-Robles MT, Pipas JM. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene. 2005;24:7729–7745. - PubMed
-
- Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, Kumar R, Jenkins RB, de Groen PC, Roche P, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat. Genet. 2004;36:744–749. - PubMed
-
- Balaban GB, Herlyn M, Clark WH, Jr, Nowell PC. Karyotypic evolution in human malignant melanoma. Cancer Genet. Cytogenet. 1986;19:113–122. - PubMed
-
- Boveri T. Concerning the origin of malignant tumours by Theodor Boveri. Harris Henry., translator. J. Cell Sci. 2008;121:1–84. and annotated. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

