Transcription leads to pervasive replisome instability in bacteria
- PMID: 28092263
- PMCID: PMC5305214
- DOI: 10.7554/eLife.19848
Transcription leads to pervasive replisome instability in bacteria
Abstract
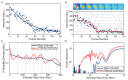
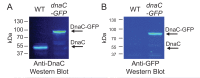
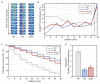
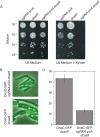
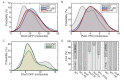
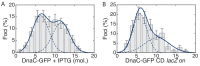
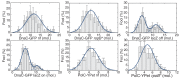
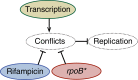
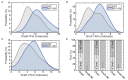
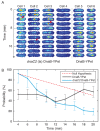
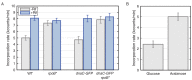
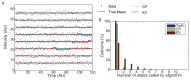
The canonical model of DNA replication describes a highly-processive and largely continuous process by which the genome is duplicated. This continuous model is based upon in vitro reconstitution and in vivo ensemble experiments. Here, we characterize the replisome-complex stoichiometry and dynamics with single-molecule resolution in bacterial cells. Strikingly, the stoichiometries of the replicative helicase, DNA polymerase, and clamp loader complexes are consistent with the presence of only one active replisome in a significant fraction of cells (>40%). Furthermore, many of the observed complexes have short lifetimes (<8 min), suggesting that replisome disassembly is quite prevalent, possibly occurring several times per cell cycle. The instability of the replisome complex is conflict-induced: transcription inhibition stabilizes these complexes, restoring the second replisome in many of the cells. Our results suggest that, in contrast to the canonical model, DNA replication is a largely discontinuous process in vivo due to pervasive replication-transcription conflicts.
Keywords: B. subtilis; DNA replication; E. coli; biophysics; cell biology; replication rates; replication-transcription conflicts; replisome; structural biology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures















References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

