Application of meta-omics techniques to understand greenhouse gas emissions originating from ruminal metabolism
- PMID: 28093073
- PMCID: PMC5240273
- DOI: 10.1186/s12711-017-0285-6
Application of meta-omics techniques to understand greenhouse gas emissions originating from ruminal metabolism
Erratum in
-
Erratum to: Application of meta-omics techniques to understand greenhouse gas emissions originating from ruminal metabolism.Genet Sel Evol. 2017 Feb 28;49(1):27. doi: 10.1186/s12711-017-0304-7. Genet Sel Evol. 2017. PMID: 28245807 Free PMC article. No abstract available.
Abstract
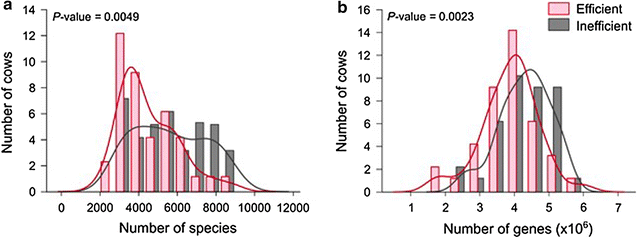
Methane emissions from ruminal fermentation contribute significantly to total anthropological greenhouse gas (GHG) emissions. New meta-omics technologies are beginning to revolutionise our understanding of the rumen microbial community structure, metabolic potential and metabolic activity. Here we explore these developments in relation to GHG emissions. Microbial rumen community analyses based on small subunit ribosomal RNA sequence analysis are not yet predictive of methane emissions from individual animals or treatments. Few metagenomics studies have been directly related to GHG emissions. In these studies, the main genes that differed in abundance between high and low methane emitters included archaeal genes involved in methanogenesis, with others that were not apparently related to methane metabolism. Unlike the taxonomic analysis up to now, the gene sets from metagenomes may have predictive value. Furthermore, metagenomic analysis predicts metabolic function better than only a taxonomic description, because different taxa share genes with the same function. Metatranscriptomics, the study of mRNA transcript abundance, should help to understand the dynamic of microbial activity rather than the gene abundance; to date, only one study has related the expression levels of methanogenic genes to methane emissions, where gene abundance failed to do so. Metaproteomics describes the proteins present in the ecosystem, and is therefore arguably a better indication of microbial metabolism. Both two-dimensional polyacrylamide gel electrophoresis and shotgun peptide sequencing methods have been used for ruminal analysis. In our unpublished studies, both methods showed an abundance of archaeal methanogenic enzymes, but neither was able to discriminate high and low emitters. Metabolomics can take several forms that appear to have predictive value for methane emissions; ruminal metabolites, milk fatty acid profiles, faecal long-chain alcohols and urinary metabolites have all shown promising results. Rumen microbial amino acid metabolism lies at the root of excessive nitrogen emissions from ruminants, yet only indirect inferences for nitrogen emissions can be drawn from meta-omics studies published so far. Annotation of meta-omics data depends on databases that are generally weak in rumen microbial entries. The Hungate 1000 project and Global Rumen Census initiatives are therefore essential to improve the interpretation of sequence/metabolic information.
Figures

References
-
- Food and Agriculture Organisation of the United Nations . Livestock’s long shadow: environmental issues and options. Rome: FAO; 2006.
-
- IPCC . Climate change 2014: synthesis report. Contribution of working groups I, II and III to the fifth assessment report of the intergovernmental panel on climate change. In: Pachauri RK, Meyer LA, editors. Core writing team. Geneva: IPCC; 2014. p. 151.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

