Genetic analysis of teat number in pigs reveals some developmental pathways independent of vertebra number and several loci which only affect a specific side
- PMID: 28093083
- PMCID: PMC5240374
- DOI: 10.1186/s12711-016-0282-1
Genetic analysis of teat number in pigs reveals some developmental pathways independent of vertebra number and several loci which only affect a specific side
Abstract
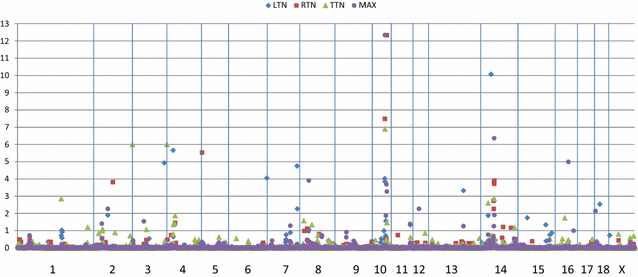
Background: Number of functional teats is an important trait in commercial swine production. As litter size increases, the number of teats must also increase to supply nutrition to all piglets. Therefore, a genome-wide association analysis was conducted to identify genomic regions that affect this trait in a commercial swine population. Genotypic data from the Illumina Porcine SNP60v1 BeadChip were available for 2951 animals with total teat number (TTN) records. A subset of these animals (n = 1828) had number of teats on each side recorded. From this information, the following traits were derived: number of teats on the left (LTN) and right side (RTN), maximum number of teats on a side (MAX), difference between LTN and RTN (L - R) and absolute value of L - R (DIF). Bayes C option of GENSEL (version 4.61) and 1-Mb windows were implemented. Identified regions that explained more than 1.5% of the genomic variation were tested in a larger group of animals (n = 5453) to estimate additive genetic effects.
Results: Marker heritabilities were highest for TTN (0.233), intermediate for individual side counts (0.088 to 0.115) and virtually nil for difference traits (0.002 for L - R and 0.006 for DIF). Each copy of the VRTN mutant allele increased teat count by 0.35 (TTN), 0.16 (LTN and RTN) and 0.19 (MAX). 15, 18, 13 and 18 one-Mb windows were detected that explained more than 1.0% of the genomic variation for TTN, LTN, RTN, and MAX, respectively. These regions cumulatively accounted for over 50% of the genomic variation of LTN, RTN and MAX, but only 30% of that of TTN. Sus scrofa chromosome SSC10:52 Mb was associated with all four count traits, while SSC10:60 and SSC14:54 Mb were associated with three count traits. Thirty-three SNPs accounted for nearly 39% of the additive genetic variation in the validation dataset. No effect of piglet sex or percentage of males in litter was detected, but birth weight was positively correlated with TTN.
Conclusions: Teat number is a heritable trait and use of genetic markers would expedite selection progress. Exploiting genetic variation associated with teat counts on each side would enhance selection focused on total teat counts. These results confirm QTL on SSC4, seven and ten and identify a novel QTL on SSC14.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

