Structural biology of supramolecular assemblies by magic-angle spinning NMR spectroscopy
- PMID: 28093096
- PMCID: PMC5483179
- DOI: 10.1017/S0033583516000159
Structural biology of supramolecular assemblies by magic-angle spinning NMR spectroscopy
Abstract
In recent years, exciting developments in instrument technology and experimental methodology have advanced the field of magic-angle spinning (MAS) nuclear magnetic resonance (NMR) to new heights. Contemporary MAS NMR yields atomic-level insights into structure and dynamics of an astounding range of biological systems, many of which cannot be studied by other methods. With the advent of fast MAS, proton detection, and novel pulse sequences, large supramolecular assemblies, such as cytoskeletal proteins and intact viruses, are now accessible for detailed analysis. In this review, we will discuss the current MAS NMR methodologies that enable characterization of complex biomolecular systems and will present examples of applications to several classes of assemblies comprising bacterial and mammalian cytoskeleton as well as human immunodeficiency virus 1 and bacteriophage viruses. The body of work reviewed herein is representative of the recent advancements in the field, with respect to the complexity of the systems studied, the quality of the data, and the significance to the biology.
Figures





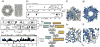





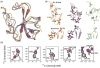
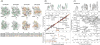
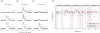








References
-
- World Health Organization. HIV/AIDS (Fact sheet No 360)
-
- ABRAMOV G, GOLDBOURT A. Nucleotide-type chemical shift assignment of the encapsulated 40 kbp dsDNA in intact bacteriophage T7 by MAS solid-state NMR. Journal of Biomolecular NMR. 2014;59(4):219–230. - PubMed
-
- ABRAMOV G, MORAG O, GOLDBOURT A. Magic-angle spinning NMR of a class I filamentous bacteriophage virus. Journal of Physical Chemistry B. 2011;115(31):9671–9680. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

