Cutting Edge: Human CD49e- NK Cells Are Tissue Resident in the Liver
- PMID: 28093522
- PMCID: PMC5296254
- DOI: 10.4049/jimmunol.1601818
Cutting Edge: Human CD49e- NK Cells Are Tissue Resident in the Liver
Abstract
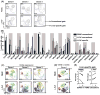
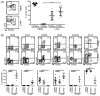
Most knowledge on NK cells is based on studies of what are now known as conventional NK cells in the mouse spleen or human peripheral blood. However, recent studies in mice indicate the presence of tissue-resident NK cells in certain organs, such as the liver, that display different markers and transcription factor dependencies as compared with conventional NK cells. In this study, we provide evidence from cytometry by time-of-flight analysis and humanized mice indicating that human CD49e- NK cells are tissue resident in the liver. Thus, these studies indicate that tissue-resident NK cells are evolutionarily conserved in humans and mice, providing a foundation to explore their role in human disease.
Copyright © 2017 by The American Association of Immunologists, Inc.
Figures

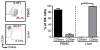
References
-
- Yokoyama WM. Chapter 17. Natural killer cells. In: Paul WE, editor. Fundamental immunology. 7. Lippincott Williams & Wilkins; Philadelphia: 2013. pp. 395–431.
-
- Sojka DK, Plougastel-Douglas B, Yang L, Pak-Wittel MA, Artyomov MN, Ivanova Y, Zhong C, Chase JM, Rothman PB, Yu J, Riley JK, Zhu J, Tian Z, Yokoyama WM. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife. 2014;3:e01659. - PMC - PubMed
-
- Daussy C, Faure F, Mayol K, Viel S, Gasteiger G, Charrier E, Bienvenu J, Henry T, Debien E, Hasan UA, Marvel J, Yoh K, Takahashi S, Prinz I, de Bernard S, Buffat L, Walzer T. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J Exp Med. 2014;211:563–577. - PMC - PubMed
-
- Seillet C, Huntington ND, Gangatirkar P, Axelsson E, Minnich M, Brady HJ, Busslinger M, Smyth MJ, Belz GT, Carotta S. Differential requirement for Nfil3 during NK cell development. J Immunol. 2014;192:2667–2676. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

