Cryptosporidium hominis gene catalog: a resource for the selection of novel Cryptosporidium vaccine candidates
- PMID: 28095366
- PMCID: PMC5070614
- DOI: 10.1093/database/baw137
Cryptosporidium hominis gene catalog: a resource for the selection of novel Cryptosporidium vaccine candidates
Abstract
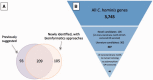
Human cryptosporidiosis, caused primarily by Cryptosporidium hominis and a subset of Cryptosporidium parvum, is a major cause of moderate-to-severe diarrhea in children under 5 years of age in developing countries and can lead to nutritional stunting and death. Cryptosporidiosis is particularly severe and potentially lethal in immunocompromised hosts. Biological and technical challenges have impeded traditional vaccinology approaches to identify novel targets for the development of vaccines against C. hominis, the predominant species associated with human disease. We deemed that the existence of genomic resources for multiple species in the genus, including a much-improved genome assembly and annotation for C. hominis, makes a reverse vaccinology approach feasible. To this end, we sought to generate a searchable online resource, termed C. hominis gene catalog, which registers all C. hominis genes and their properties relevant for the identification and prioritization of candidate vaccine antigens, including physical attributes, properties related to antigenic potential and expression data. Using bioinformatic approaches, we identified ∼400 C. hominis genes containing properties typical of surface-exposed antigens, such as predicted glycosylphosphatidylinositol (GPI)-anchor motifs, multiple transmembrane motifs and/or signal peptides targeting the encoded protein to the secretory pathway. This set can be narrowed further, e.g. by focusing on potential GPI-anchored proteins lacking homologs in the human genome, but with homologs in the other Cryptosporidium species for which genomic data are available, and with low amino acid polymorphism. Additional selection criteria related to recombinant expression and purification include minimizing predicted post-translation modifications and potential disulfide bonds. Forty proteins satisfying these criteria were selected from 3745 proteins in the updated C. hominis annotation. The immunogenic potential of a few of these is currently being tested.Database URL: http://cryptogc.igs.umaryland.edu.
© The Author(s) 2016. Published by Oxford University Press.
Figures



References
-
- Liu L., Oza S., Hogan D. et al. (2015) Global, regional, and national causes of child mortality in 2000-13, with projections to inform post-2015 priorities: an updated systematic analysis. Lancet, 385, 430–440. - PubMed
-
- Kotloff K.L., Nataro J.P., Blackwelder W.C. et al. (2013) Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet, 382, 209–222. - PubMed
-
- Xiao L., Bern C., Limor J. et al. (2001) Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J. Infect. Dis., 183, 492–497. - PubMed
-
- Cama V.A., Bern C., Sulaiman I.M. et al. (2003) Cryptosporidium species and genotypes in HIV-positive patients in Lima, Peru. J. Eukaryot. Microbiol., 50 Suppl, 531–533. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

