Replication-Coupled Recruitment of Viral and Cellular Factors to Herpes Simplex Virus Type 1 Replication Forks for the Maintenance and Expression of Viral Genomes
- PMID: 28095497
- PMCID: PMC5271410
- DOI: 10.1371/journal.ppat.1006166
Replication-Coupled Recruitment of Viral and Cellular Factors to Herpes Simplex Virus Type 1 Replication Forks for the Maintenance and Expression of Viral Genomes
Abstract
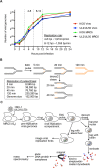
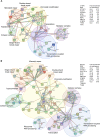
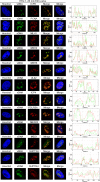
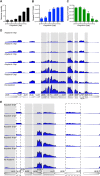
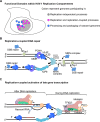
Herpes simplex virus type 1 (HSV-1) infects over half the human population. Much of the infectious cycle occurs in the nucleus of cells where the virus has evolved mechanisms to manipulate host processes for the production of virus. The genome of HSV-1 is coordinately expressed, maintained, and replicated such that progeny virions are produced within 4-6 hours post infection. In this study, we selectively purify HSV-1 replication forks and associated proteins from virus-infected cells and identify select viral and cellular replication, repair, and transcription factors that associate with viral replication forks. Pulse chase analyses and imaging studies reveal temporal and spatial dynamics between viral replication forks and associated proteins and demonstrate that several DNA repair complexes and key transcription factors are recruited to or near replication forks. Consistent with these observations we show that the initiation of viral DNA replication is sufficient to license late gene transcription. These data provide insight into mechanisms that couple HSV-1 DNA replication with transcription and repair for the coordinated expression and maintenance of the viral genome.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- McGeoch DJ, Dalrymple MA, Davison AJ, Dolan A, Frame MC, et al. (1988) The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol 69 (Pt 7): 1531–1574. - PubMed
-
- McGeoch DJ, Dolan A, Donald S, Rixon FJ (1985) Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J Mol Biol 181: 1–13. - PubMed
-
- Alwine JC, Steinhart WL, Hill CW (1974) Transcription of herpes simplex type 1 DNA in nuclei isolated from infected HEp-2 and KB cells. Virology 60: 302–307. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

