The pangenome of (Antarctic) Pseudoalteromonas bacteria: evolutionary and functional insights
- PMID: 28095778
- PMCID: PMC5240218
- DOI: 10.1186/s12864-016-3382-y
The pangenome of (Antarctic) Pseudoalteromonas bacteria: evolutionary and functional insights
Abstract
Background: Pseudoalteromonas is a genus of ubiquitous marine bacteria used as model organisms to study the biological mechanisms involved in the adaptation to cold conditions. A remarkable feature shared by these bacteria is their ability to produce secondary metabolites with a strong antimicrobial and antitumor activity. Despite their biotechnological relevance, representatives of this genus are still lacking (with few exceptions) an extensive genomic characterization, including features involved in the evolution of secondary metabolites production. Indeed, biotechnological applications would greatly benefit from such analysis.
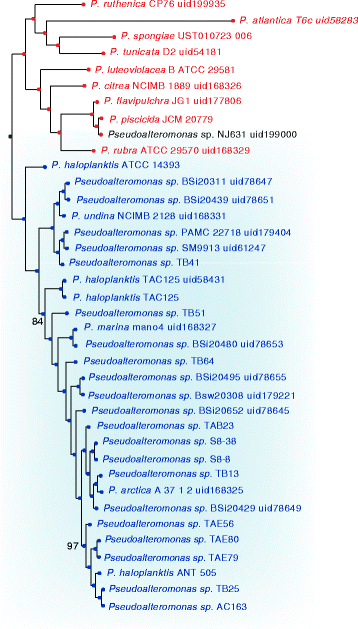
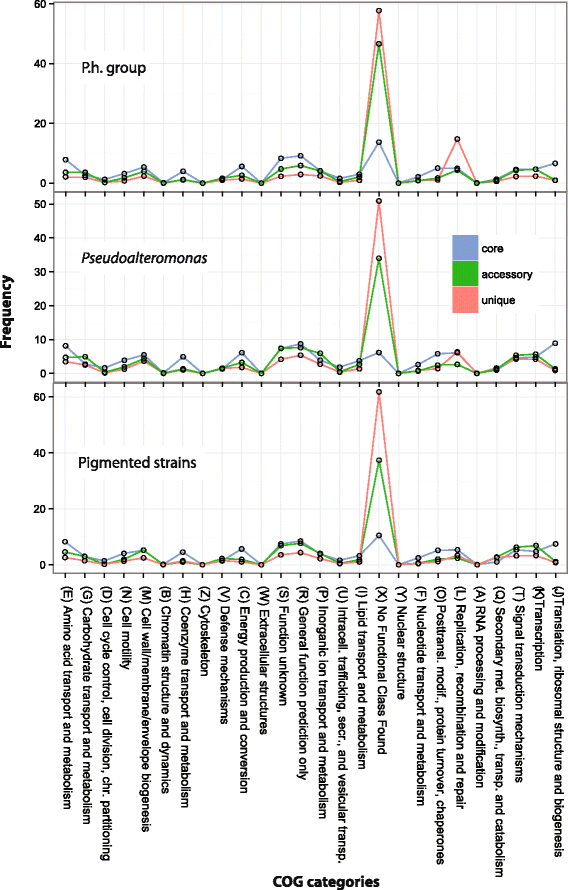
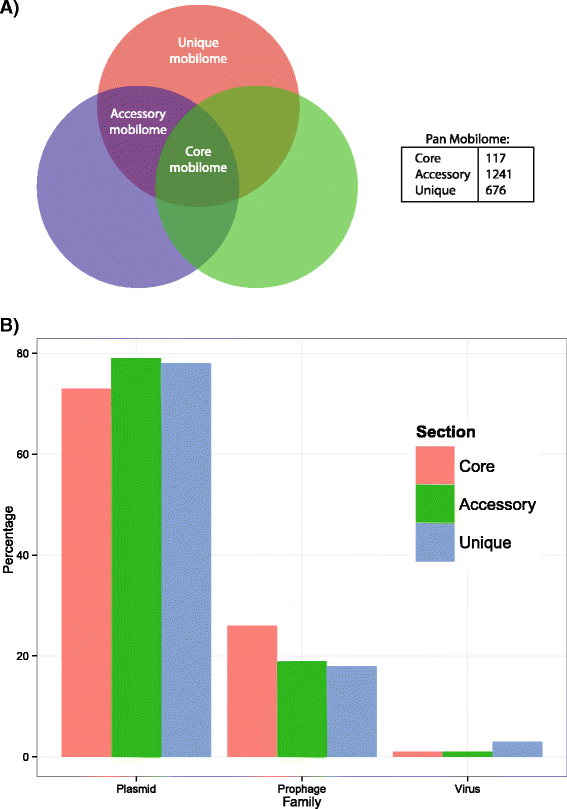
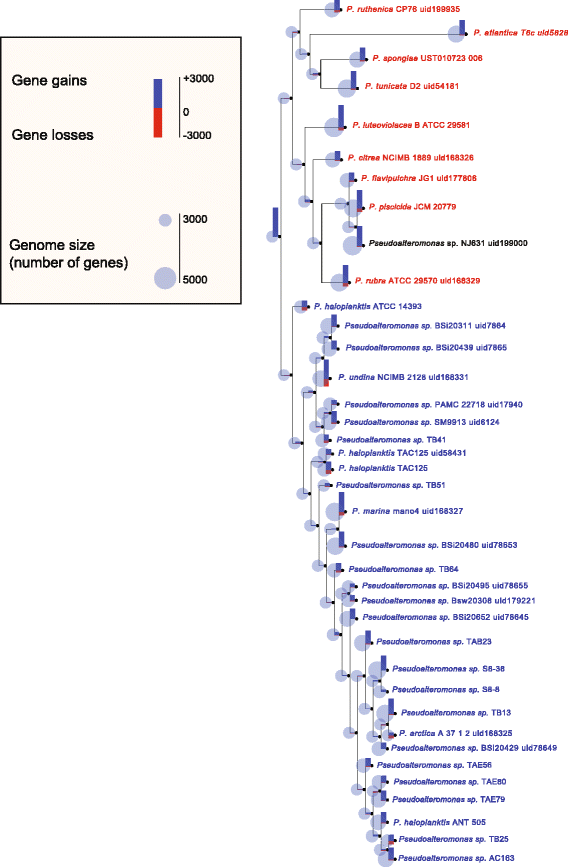
Results: Here, we analyzed the genomes of 38 strains belonging to different Pseudoalteromonas species and isolated from diverse ecological niches, including extreme ones (i.e. Antarctica). These sequences were used to reconstruct the largest Pseudoalteromonas pangenome computed so far, including also the two main groups of Pseudoalteromonas strains (pigmented and not pigmented strains). The downstream analyses were conducted to describe the genomic diversity, both at genus and group levels. This allowed highlighting a remarkable genomic heterogeneity, even for closely related strains. We drafted all the main evolutionary steps that led to the current structure and gene content of Pseudoalteromonas representatives. These, most likely, included an extensive genome reduction and a strong contribution of Horizontal Gene Transfer (HGT), which affected biotechnologically relevant gene sets and occurred in a strain-specific fashion. Furthermore, this study also identified the genomic determinants related to some of the most interesting features of the Pseudoalteromonas representatives, such as the production of secondary metabolites, the adaptation to cold temperatures and the resistance to abiotic compounds.
Conclusions: This study poses the bases for a comprehensive understanding of the evolutionary trajectories followed in time by this peculiar bacterial genus and for a focused exploitation of their biotechnological potential.
Keywords: Antarctic bacteria; Antibiotics; Comparative genomics; Horizontal gene transfer; Marine bacteria; Microbial evolution; Pangenome; Pseudoalteromonas.
Figures


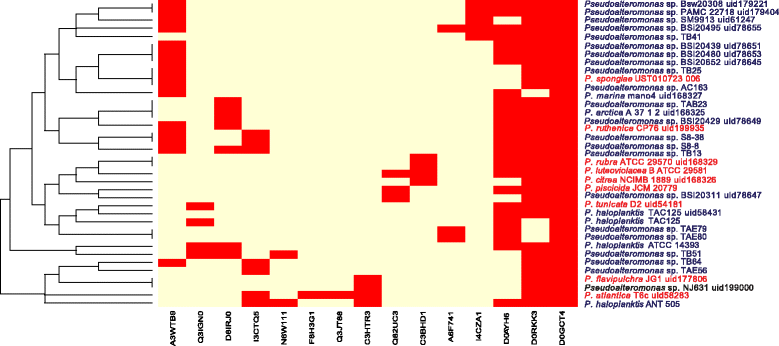
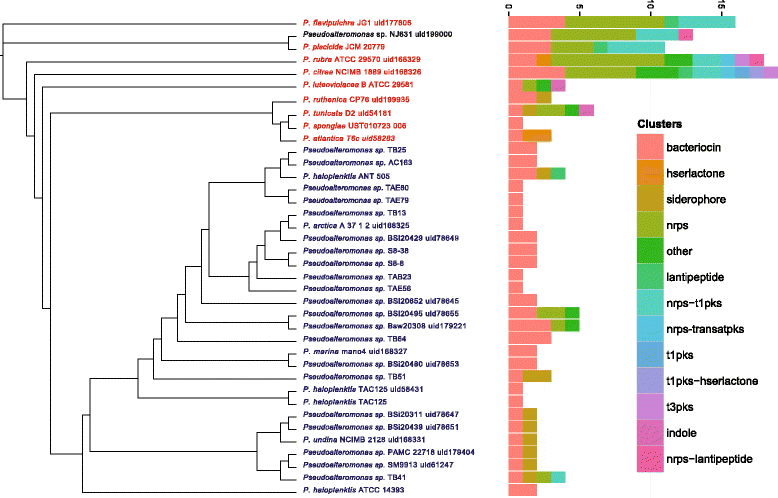
References
-
- Ivanova EP, Flavier S, Christen R. Phylogenetic relationships among marine Alteromonas-like proteobacteria: emended description of the family Alteromonadaceae and proposal of Pseudoalteromonadaceae fam. nov., Colwelliaceae fam. nov., Shewanellaceae fam. nov., Moritellaceae fam. nov., Ferrimonadaceae fam. nov., Idiomarinaceae fam. nov. and Psychromonadaceae fam. nov. Int J Syst Evol Microbiol. 2004;54(Pt 5):1773–88. doi: 10.1099/ijs.0.02997-0. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

