Toward the Atomic Structure of PrPSc
- PMID: 28096267
- PMCID: PMC5585850
- DOI: 10.1101/cshperspect.a031336
Toward the Atomic Structure of PrPSc
Abstract
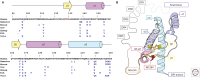
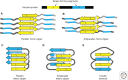
In this review, we detail our current knowledge of PrPSc structure on the basis of structural and computational studies. We discuss the progress toward an atomic resolution description of PrPSc and results from the broader field of amyloid studies that may further inform our knowledge of this structure. Moreover, we summarize work that investigates the role of PrPSc structure in its toxicity, transmissibility, and species specificity. We look forward to an atomic model of PrPSc, which is expected to bring diagnostics and/or therapeutics to the field of prion disease.
Copyright © 2017 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
