The NBS-LRR architectures of plant R-proteins and metazoan NLRs evolved in independent events
- PMID: 28096345
- PMCID: PMC5293065
- DOI: 10.1073/pnas.1619730114
The NBS-LRR architectures of plant R-proteins and metazoan NLRs evolved in independent events
Abstract
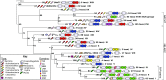
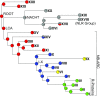
There are intriguing parallels between plants and animals, with respect to the structures of their innate immune receptors, that suggest universal principles of innate immunity. The cytosolic nucleotide binding site-leucine rich repeat (NBS-LRR) resistance proteins of plants (R-proteins) and the so-called NOD-like receptors of animals (NLRs) share a domain architecture that includes a STAND (signal transduction ATPases with numerous domains) family NTPase followed by a series of LRRs, suggesting inheritance from a common ancestor with that architecture. Focusing on the STAND NTPases of plant R-proteins, animal NLRs, and their homologs that represent the NB-ARC (nucleotide-binding adaptor shared by APAF-1, certain R gene products and CED-4) and NACHT (named for NAIP, CIIA, HET-E, and TEP1) subfamilies of the STAND NTPases, we analyzed the phylogenetic distribution of the NBS-LRR domain architecture, used maximum-likelihood methods to infer a phylogeny of the NTPase domains of R-proteins, and reconstructed the domain structure of the protein containing the common ancestor of the STAND NTPase domain of R-proteins and NLRs. Our analyses reject monophyly of plant R-proteins and NLRs and suggest that the protein containing the last common ancestor of the STAND NTPases of plant R-proteins and animal NLRs (and, by extension, all NB-ARC and NACHT domains) possessed a domain structure that included a STAND NTPase paired with a series of tetratricopeptide repeats. These analyses reject the hypothesis that the domain architecture of R-proteins and NLRs was inherited from a common ancestor and instead suggest the domain architecture evolved at least twice. It remains unclear whether the NBS-LRR architectures were innovations of plants and animals themselves or were acquired by one or both lineages through horizontal gene transfer.
Keywords: NLR; NOD-like receptors; R-protein; evolution; innate immunity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

