Establishment of expression-state boundaries by Rif1 and Taz1 in fission yeast
- PMID: 28096402
- PMCID: PMC5293076
- DOI: 10.1073/pnas.1614837114
Establishment of expression-state boundaries by Rif1 and Taz1 in fission yeast
Abstract
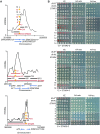
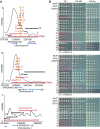
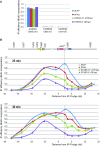
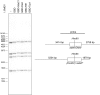
The Shelterin component Rif1 has emerged as a global regulator of the replication-timing program in all eukaryotes examined to date, possibly by modulating the 3D-organization of the genome. In fission yeast a second Shelterin component, Taz1, might share similar functions. Here, we identified unexpected properties for Rif1 and Taz1 by conducting high-throughput genetic screens designed to identify cis- and trans-acting factors capable of creating heterochromatin-euchromatin boundaries in fission yeast. The preponderance of cis-acting elements identified in the screens originated from genomic loci bound by Taz1 and associated with origins of replication whose firing is repressed by Taz1 and Rif1. Boundary formation and gene silencing by these elements required Taz1 and Rif1 and coincided with altered replication timing in the region. Thus, small chromosomal elements sensitive to Taz1 and Rif1 (STAR) could simultaneously regulate gene expression and DNA replication over a large domain, at the edge of which they established a heterochromatin-euchromatin boundary. Taz1, Rif1, and Rif1-associated protein phosphatases Sds21 and Dis2 were each sufficient to establish a boundary when tethered to DNA. Moreover, efficient boundary formation required the amino-terminal domain of the Mcm4 replicative helicase onto which the antagonistic activities of the replication-promoting Dbf4-dependent kinase and Rif1-recruited phosphatases are believed to converge to control replication origin firing. Altogether these observations provide an insight into a coordinated control of DNA replication and organization of the genome into expression domains.
Keywords: DNA replication program; chromatin boundaries; fission yeast; gene silencing; heterochromatin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Barkess G, West AG. Chromatin insulator elements: Establishing barriers to set heterochromatin boundaries. Epigenomics. 2012;4(1):67–80. - PubMed
-
- Ali T, Renkawitz R, Bartkuhn M. Insulators and domains of gene expression. Curr Opin Genet Dev. 2016;37:17–26. - PubMed
-
- Keller C, Kulasegaran-Shylini R, Shimada Y, Hotz HR, Bühler M. Noncoding RNAs prevent spreading of a repressive histone mark. Nat Struct Mol Biol. 2013;20(8):994–1000. - PubMed
-
- Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64(5):941–950. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

