Comprehensive transcriptome analysis provides new insights into nutritional strategies and phylogenetic relationships of chrysophytes
- PMID: 28097055
- PMCID: PMC5228505
- DOI: 10.7717/peerj.2832
Comprehensive transcriptome analysis provides new insights into nutritional strategies and phylogenetic relationships of chrysophytes
Abstract
Background: Chrysophytes are protist model species in ecology and ecophysiology and important grazers of bacteria-sized microorganisms and primary producers. However, they have not yet been investigated in detail at the molecular level, and no genomic and only little transcriptomic information is available. Chrysophytes exhibit different trophic modes: while phototrophic chrysophytes perform only photosynthesis, mixotrophs can gain carbon from bacterial food as well as from photosynthesis, and heterotrophs solely feed on bacteria-sized microorganisms. Recent phylogenies and megasystematics demonstrate an immense complexity of eukaryotic diversity with numerous transitions between phototrophic and heterotrophic organisms. The question we aim to answer is how the diverse nutritional strategies, accompanied or brought about by a reduction of the plasmid and size reduction in heterotrophic strains, affect physiology and molecular processes.
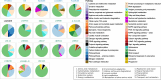
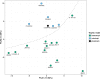
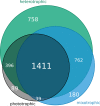
Results: We sequenced the mRNA of 18 chrysophyte strains on the Illumina HiSeq platform and analysed the transcriptomes to determine relations between the trophic mode (mixotrophic vs. heterotrophic) and gene expression. We observed an enrichment of genes for photosynthesis, porphyrin and chlorophyll metabolism for phototrophic and mixotrophic strains that can perform photosynthesis. Genes involved in nutrient absorption, environmental information processing and various transporters (e.g., monosaccharide, peptide, lipid transporters) were present or highly expressed only in heterotrophic strains that have to sense, digest and absorb bacterial food. We furthermore present a transcriptome-based alignment-free phylogeny construction approach using transcripts assembled from short reads to determine the evolutionary relationships between the strains and the possible influence of nutritional strategies on the reconstructed phylogeny. We discuss the resulting phylogenies in comparison to those from established approaches based on ribosomal RNA and orthologous genes. Finally, we make functionally annotated reference transcriptomes of each strain available to the community, significantly enhancing publicly available data on Chrysophyceae.
Conclusions: Our study is the first comprehensive transcriptomic characterisation of a diverse set of Chrysophyceaen strains. In addition, we showcase the possibility of inferring phylogenies from assembled transcriptomes using an alignment-free approach. The raw and functionally annotated data we provide will prove beneficial for further examination of the diversity within this taxon. Our molecular characterisation of different trophic modes presents a first such example.
Keywords: Alignment-free phylogeny; Chrysophyceae; Differential expression analysis; Molecular phylogeny; Nutritional strategy; Pathway analysis; Protists; RNA-Seq; Transcriptomics.
Conflict of interest statement
Sven Rahmann is an Academic Editor for PeerJ.
Figures






References
-
- Adl SM, Simpson AGB, Lane CE, Lukeš J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, Heiss A, Hoppenrath M, Lara E, Le Gall L, Lynn DH, McManus H, Mitchell EAD, Mozley-Stanridge SE, Parfrey LW, Pawlowski J, Rueckert S, Shadwick RS, Shadwick L, Schoch CL, Smirnov A, Spiegel FW. The revised classification of eukaryotes. The Journal of Eukaryotic Microbiology. 2012;59(5):429–493. doi: 10.1111/j.1550-7408.2012.00644.x. - DOI - PMC - PubMed
-
- Ahmed N. Metabolism and functions of polyamines. Biochemical Education. 1987;15(3):106–110. doi: 10.1016/0307-4412(87)90037-9. - DOI
-
- Andersen RA. Molecular systematics of the Chrysophyceae and Synurophyceae. In: Brodie J, Lewis J, editors. Unravelling the algae —the past, present, and future of algal systematics. CRC Press; Boca Raton: 2007. pp. 285–313. (Systematics association special volumes).
-
- Andrews S. FastQC a quality control tool for high throughput sequence data. [Software] 2012. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
-
- Beisser D, Graupner N, Bock C, Wodniok S, Grossmann L, Vos M, Sures B, Rahmann S, Boenigk J. Raw sequence data and assembled transcripts at ENA EBI. 2016. http://www.ebi.ac.uk/ena/data/search?query=PRJEB13662. [2016 July 25]. http://www.ebi.ac.uk/ena/data/search?query=PRJEB13662
LinkOut - more resources
Full Text Sources
Other Literature Sources

