A comparative study of the degradation of yeast cyclins Cln1 and Cln2
- PMID: 28097090
- PMCID: PMC5221467
- DOI: 10.1002/2211-5463.12157
A comparative study of the degradation of yeast cyclins Cln1 and Cln2
Abstract
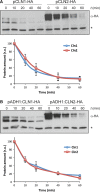
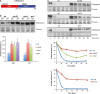
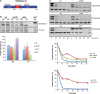
The yeast cyclins Cln1 and Cln2 are very similar in both sequence and function, but some differences in their functionality and localization have been recently described. The control of Cln1 and Cln2 cellular levels is crucial for proper cell cycle initiation. In this work, we analyzed the degradation patterns of Cln1 and Cln2 in order to further investigate the possible differences between them. Both cyclins show the same half-life but, while Cln2 degradation depends on ubiquitin ligases SCFGrr1 and SCFCdc4, Cln1 is affected only by SCFGrr1. Degradation analysis of chimeric cyclins, constructed by combining fragments from Cln1 and Cln2, identifies the N-terminal sequence of the proteins as responsible of the cyclin degradation pattern. In particular, the N-terminal region of Cln2 is required to mediate degradation by SCFCdc4. This region is involved in nuclear import of Cln1 and Cln2, which suggests that differences in degradation may be due to differences in localization. Moreover, a comparison of the cyclins that differ only in the presence of the Cln2 nuclear export signal indicates a greater instability of exported cyclins, thus reinforcing the idea that cyclin stability is influenced by their localization.
Keywords: Cln1; Cln2; SCF ubiquitin ligase; Saccharomyces cerevisiae; cell cycle; cyclin.
Figures





References
-
- Andrews B and Measday V (1998) The cyclin family of budding yeast: abundant use of a good idea. Trends Genet 14, 66–72. - PubMed
-
- Breeden LL (2003) Periodic transcription: a cycle within a cycle. Curr Biol 13, R31–R38. - PubMed
-
- McInerny CJ (2011) Cell cycle regulated gene expression in yeasts. Adv Genet 73, 51–85. - PubMed
-
- Simon I, Barnett J, Hannett N, Harbison CT, Rinaldi NJ, Volkert TL, Wyrick JJ, Zeitlinger J, Gifford DK, Jaakkola TS et al (2001) Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106, 697–708. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

