Exome and genome sequencing of nasopharynx cancer identifies NF-κB pathway activating mutations
- PMID: 28098136
- PMCID: PMC5253631
- DOI: 10.1038/ncomms14121
Exome and genome sequencing of nasopharynx cancer identifies NF-κB pathway activating mutations
Abstract
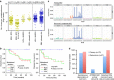
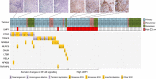
Nasopharyngeal carcinoma (NPC) is an aggressive head and neck cancer characterized by Epstein-Barr virus (EBV) infection and dense lymphocyte infiltration. The scarcity of NPC genomic data hinders the understanding of NPC biology, disease progression and rational therapy design. Here we performed whole-exome sequencing (WES) on 111 micro-dissected EBV-positive NPCs, with 15 cases subjected to further whole-genome sequencing (WGS), to determine its mutational landscape. We identified enrichment for genomic aberrations of multiple negative regulators of the NF-κB pathway, including CYLD, TRAF3, NFKBIA and NLRC5, in a total of 41% of cases. Functional analysis confirmed inactivating CYLD mutations as drivers for NPC cell growth. The EBV oncoprotein latent membrane protein 1 (LMP1) functions to constitutively activate NF-κB signalling, and we observed mutual exclusivity among tumours with somatic NF-κB pathway aberrations and LMP1-overexpression, suggesting that NF-κB activation is selected for by both somatic and viral events during NPC pathogenesis.
Conflict of interest statement
V.W.Y. serves as a scientific consultant for Novartis, Hong Kong. B.B.Y.M. received research grant and serves the advisory board from Novartis, Hong Kong.
Figures





References
-
- Bruce J. P., Yip K., Bratman S. V., Ito E. & Liu F. F. Nasopharyngeal cancer: molecular landscape. J. Clin. Oncol. 33, 3346–3355 (2015). - PubMed
-
- Chua M. L., Wee J. T., Hui E. P. & Chan A. T. Nasopharyngeal carcinoma. Lancet 387, 1012–1024 (2016). - PubMed
-
- Lin D. C. et al. The genomic landscape of nasopharyngeal carcinoma. Nat. Genet. 46, 866–871 (2014). - PubMed
-
- Lo K. W., To K. F. & Huang D. P. Focus on nasopharyngeal carcinoma. Cancer Cell 5, 423–428 (2004). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

