Epigenetic modifications at DMRs of placental genes are subjected to variations in normal gestation, pathological conditions and folate supplementation
- PMID: 28098215
- PMCID: PMC5241688
- DOI: 10.1038/srep40774
Epigenetic modifications at DMRs of placental genes are subjected to variations in normal gestation, pathological conditions and folate supplementation
Abstract
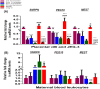
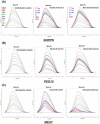
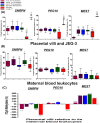
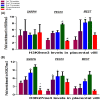
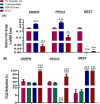
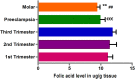
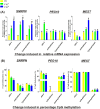
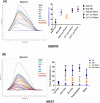
Invasive placentation and cancer development shares many similar molecular and epigenetic pathways. Paternally expressed, growth promoting genes (SNRPN, PEG10 and MEST) which are known to play crucial role in tumorogenesis, are not well studied during placentation. This study reports for the first time of the impact of gestational-age, pathological conditions and folic acid supplementation on dynamic nature of DNA and histone methylation present at their differentially methylated regions (DMRs). Here, we reported the association between low DNA methylation/H3K27me3 and higher expression of SNRPN, PEG10 and MEST in highly proliferating normal early gestational placenta. Molar and preeclamptic placental villi, exhibited aberrant changes in methylation levels at DMRs of these genes, leading to higher and lower expression of these genes, respectively, in reference to their respective control groups. Moreover, folate supplementation could induce gene specific changes in mRNA expression in placental cell lines. Further, MEST and SNRPN DMRs were observed to show the potential to act as novel fetal DNA markers in maternal plasma. Thus, variation in methylation levels at these DMRs regulate normal placentation and placental disorders. Additionally, the methylation at these DMRs might also be susceptible to folic acid supplementation and has the potential to be utilized in clinical diagnosis.
Figures
References
-
- Ferretti C., Bruni L., Dangles-Marie V., Pecking A. P. & Bellet D. Molecular circuits shared by placental and cancer cells, and their implications in the proliferative, invasive and migratory capacities of trophoblasts. Hum Reprod Update 13, 121–141, doi: dml048 10.1093/humupd/dml048, doi: 10.1038/srep02696(2013) J. Cell Biol (2007). - DOI - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

