IsomiR expression patterns in canonical and Dicer‑independent microRNAs
- PMID: 28098889
- PMCID: PMC5367367
- DOI: 10.3892/mmr.2017.6117
IsomiR expression patterns in canonical and Dicer‑independent microRNAs
Abstract
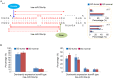
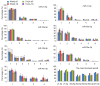
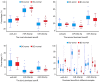
Multiple microRNA (miRNA) variants, known as isomiRs, are extensively distributed in miRNA loci and predominantly derive from the alternative cleavage of Drosha/Dicer and 3'addition events. The present study aimed to investigate the expression patterns of multiple isomiRs in typical miRNA and Dicer‑independent miRNA loci by conducting evolutionary and expression analysis using public datasets. Although different miRNA maturation processes exist, multiple isomiRs can be detected by similar expression distributions. However, isomiR expression in Dicer‑independent miRNA loci tends to be at a moderate level, particularly for random distribution in the ends that are split by Dicer in the typical miRNA loci. Compared with the mature miRNA locus (dominant miRNA locus), the non‑dominant miRNA locus indicates an expression distribution similar to that of the Dicer‑independent miRNA locus. These results increase the understanding of multiple isomiRs in the progression of diseases.
Figures


Similar articles
-
A challenge for miRNA: multiple isomiRs in miRNAomics.Gene. 2014 Jul 1;544(1):1-7. doi: 10.1016/j.gene.2014.04.039. Epub 2014 Apr 21. Gene. 2014. PMID: 24768184 Review.
-
Close association between paralogous multiple isomiRs and paralogous/orthologues miRNA sequences implicates dominant sequence selection across various animal species.Gene. 2013 Sep 25;527(2):624-9. doi: 10.1016/j.gene.2013.06.083. Epub 2013 Jul 13. Gene. 2013. PMID: 23856130
-
Evolutionary and expression analysis of miR-#-5p and miR-#-3p at the miRNAs/isomiRs levels.Biomed Res Int. 2015;2015:168358. doi: 10.1155/2015/168358. Epub 2015 May 5. Biomed Res Int. 2015. PMID: 26075215 Free PMC article.
-
A genome-wide screen for non-template nucleotides and isomiR repertoires in miRNAs indicates dynamic and versatile microRNAome.Mol Biol Rep. 2014 Oct;41(10):6649-58. doi: 10.1007/s11033-014-3548-0. Epub 2014 Jul 4. Mol Biol Rep. 2014. PMID: 24993117
-
IsomiRs have functional importance.Malays J Pathol. 2015 Aug;37(2):73-81. Malays J Pathol. 2015. PMID: 26277662 Review.
Cited by
-
Identifying High Confidence microRNAs in the Developing Seeds of Jatropha curcas.Sci Rep. 2019 Mar 14;9(1):4510. doi: 10.1038/s41598-019-41189-y. Sci Rep. 2019. PMID: 30872797 Free PMC article.
-
5'‑isomiR is the most abundant sequence of miR‑1246, a candidate biomarker of lung cancer, in serum.Mol Med Rep. 2023 Apr;27(4):92. doi: 10.3892/mmr.2023.12979. Epub 2023 Mar 24. Mol Med Rep. 2023. PMID: 36960865 Free PMC article.
-
A Novel Tissue Atlas and Online Tool for the Interrogation of Small RNA Expression in Human Tissues and Biofluids.Front Cell Dev Biol. 2022 Mar 4;10:804164. doi: 10.3389/fcell.2022.804164. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35317387 Free PMC article.
-
Interactions of host miRNAs in the flavivirus 3´UTR genome: From bioinformatics predictions to practical approaches.Front Cell Infect Microbiol. 2022 Oct 13;12:976843. doi: 10.3389/fcimb.2022.976843. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36310869 Free PMC article. Review.
-
Novel and Haplotype Specific MicroRNAs Encoded by the Major Histocompatibility Complex.Sci Rep. 2018 Mar 1;8(1):3832. doi: 10.1038/s41598-018-19427-6. Sci Rep. 2018. PMID: 29497078 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

