Mitochondrial respiratory gene expression is suppressed in many cancers
- PMID: 28099114
- PMCID: PMC5243113
- DOI: 10.7554/eLife.21592
Mitochondrial respiratory gene expression is suppressed in many cancers
Abstract
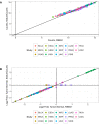
The fundamental metabolic decision of a cell, the balance between respiration and fermentation, rests in part on expression of the mitochondrial genome (mtDNA) and coordination with expression of the nuclear genome (nuDNA). Previously we described mtDNA copy number depletion across many solid tumor types (Reznik et al., 2016). Here, we use orthogonal RNA-sequencing data to quantify mtDNA expression (mtRNA), and report analogously lower expression of mtRNA in tumors (relative to normal tissue) across a majority of cancer types. Several cancers exhibit a trio of mutually consistent evidence suggesting a drop in respiratory activity: depletion of mtDNA copy number, decreases in mtRNA levels, and decreases in expression of nuDNA-encoded respiratory proteins. Intriguingly, a minority of cancer types exhibit a drop in mtDNA expression but an increase in nuDNA expression of respiratory proteins, with unknown implications for respiratory activity. Our results indicate suppression of respiratory gene expression across many cancer types.
Keywords: cancer; cancer biology; computational biology; human; metabolism; mitochondria; mtDNA; mtRNA; respiration; systems biology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures










References
-
- Cancer Genome Atlas Research Network. Abeshouse A, Ahn J, Akbani R, Ally A, Amin S, Andry CD, Annala M, Aprikian A, Armenia J, Arora A, Auman JT, Balasundaram M, Balu S, Barbieri CE, Bauer T, Benz CC, Bergeron A, Beroukhim R, Berrios M, Bivol A, Bodenheimer T, Boice L, Bootwalla MS, Borges dos Reis R, Boutros PC, Bowen J, Bowlby R, Boyd J, Bradley RK, Breggia A, Brimo F, Bristow CA, Brooks D, Broom BM, Bryce AH, Bubley G, Burks E, Butterfield YS, Button M, Canes D, Carlotti CG, Carlsen R, Carmel M, Carroll PR, Carter SL, Cartun R, Carver BS, Chan JM, Chang MT, Chen Y, Cherniack AD, Chevalier S, Chin L, Cho J, Chu A, Chuah E, Chudamani S, Cibulskis K, Ciriello G, Clarke A, Cooperberg MR, Corcoran NM, Costello AJ, Cowan J, Crain D, Curley E, David K, Demchok JA, Demichelis F, Dhalla N, Dhir R, Doueik A, Drake B, Dvinge H, Dyakova N, Felau I, Ferguson ML, Frazer S, Freedland S, Fu Y, Gabriel SB, Gao J, Gardner J, Gastier-Foster JM, Gehlenborg N, Gerken M, Gerstein MB, Getz G, Godwin AK, Gopalan A, Graefen M, Graim K, Gribbin T, Guin R, Gupta M, Hadjipanayis A, Haider S, Hamel L, Hayes DN, Heiman DI, Hess J, Hoadley KA, Holbrook AH, Holt RA, Holway A, Hovens CM, Hoyle AP, Huang M, Hutter CM, Ittmann M, Iype L, Jefferys SR, Jones CD, Jones SJ, Juhl H, Kahles A, Kane CJ, Kasaian K, Kerger M, Khurana E, Kim J, Klein RJ, Kucherlapati R, Lacombe L, Ladanyi M, Lai PH, Laird PW, Lander ES, Latour M, Lawrence MS, Lau K, LeBien T, Lee D, Lee S, Lehmann KV, Leraas KM, Leshchiner I, Leung R, Libertino JA, Lichtenberg TM, Lin P, Linehan WM, Ling S, Lippman SM, Liu J, Liu W, Lochovsky L, Loda M, Logothetis C, Lolla L, Longacre T, Lu Y, Luo J, Ma Y, Mahadeshwar HS, Mallery D, Mariamidze A, Marra MA, Mayo M, McCall S, McKercher G, Meng S, Mes-Masson AM, Merino MJ, Meyerson M, Mieczkowski PA, Mills GB, Mills Shaw KR, Minner S, Moinzadeh A, Moore RA, Morris S, Morrison C, Mose LE, Mungall AJ, Murray BA, Myers JB, Naresh R, Nelson J, Nelson MA, Nelson PS, Newton Y, Noble MS, Noushmehr H, Nykter M, Pantazi A, Parfenov M, Park PJ, Parker JS, Paulauskis J, Penny R, Perou CM, Piché A, Pihl T, Pinto PA, Prandi D, Protopopov A, Ramirez NC, Rao A, Rathmell WK, Rätsch G, Ren X, Reuter VE, Reynolds SM, Rhie SK, Rieger-Christ K, Roach J, Robertson AG, Robinson B, Rubin MA, Saad F, Sadeghi S, Saksena G, Saller C, Salner A, Sanchez-Vega F, Sander C, Sandusky G, Sauter G, Sboner A, Scardino PT, Scarlata E, Schein JE, Schlomm T, Schmidt LS, Schultz N, Schumacher SE, Seidman J, Neder L, Seth S, Sharp A, Shelton C, Shelton T, Shen H, Shen R, Sherman M, Sheth M, Shi Y, Shih J, Shmulevich I, Simko J, Simon R, Simons JV, Sipahimalani P, Skelly T, Sofia HJ, Soloway MG, Song X, Sorcini A, Sougnez C, Stepa S, Stewart C, Stewart J, Stuart JM, Sullivan TB, Sun C, Sun H, Tam A, Tan D, Tang J, Tarnuzzer R, Tarvin K, Taylor BS, Teebagy P, Tenggara I, Têtu B, Tewari A, Thiessen N, Thompson T, Thorne LB, Tirapelli DP, Tomlins SA, Trevisan FA, Troncoso P, True LD, Tsourlakis MC, Tyekucheva S, Van Allen E, Van Den Berg DJ, Veluvolu U, Verhaak R, Vocke CD, Voet D, Wan Y, Wang Q, Wang W, Wang Z, Weinhold N, Weinstein JN, Weisenberger DJ, Wilkerson MD, Wise L, Witte J, Zhang J, Zhang J, Zhang W, Yang L, Zhang J. The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

