Genomic deletion of malic enzyme 2 confers collateral lethality in pancreatic cancer
- PMID: 28099419
- PMCID: PMC5398413
- DOI: 10.1038/nature21052
Genomic deletion of malic enzyme 2 confers collateral lethality in pancreatic cancer
Abstract
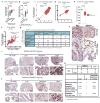
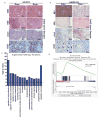
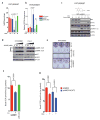
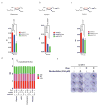
The genome of pancreatic ductal adenocarcinoma (PDAC) frequently contains deletions of tumour suppressor gene loci, most notably SMAD4, which is homozygously deleted in nearly one-third of cases. As loss of neighbouring housekeeping genes can confer collateral lethality, we sought to determine whether loss of the metabolic gene malic enzyme 2 (ME2) in the SMAD4 locus would create cancer-specific metabolic vulnerability upon targeting of its paralogous isoform ME3. The mitochondrial malic enzymes (ME2 and ME3) are oxidative decarboxylases that catalyse the conversion of malate to pyruvate and are essential for NADPH regeneration and reactive oxygen species homeostasis. Here we show that ME3 depletion selectively kills ME2-null PDAC cells in a manner consistent with an essential function for ME3 in ME2-null cancer cells. Mechanistically, integrated metabolomic and molecular investigation of cells deficient in mitochondrial malic enzymes revealed diminished NADPH production and consequent high levels of reactive oxygen species. These changes activate AMP activated protein kinase (AMPK), which in turn directly suppresses sterol regulatory element-binding protein 1 (SREBP1)-directed transcription of its direct targets including the BCAT2 branched-chain amino acid transaminase 2) gene. BCAT2 catalyses the transfer of the amino group from branched-chain amino acids to α-ketoglutarate (α-KG) thereby regenerating glutamate, which functions in part to support de novo nucleotide synthesis. Thus, mitochondrial malic enzyme deficiency, which results in impaired NADPH production, provides a prime 'collateral lethality' therapeutic strategy for the treatment of a substantial fraction of patients diagnosed with this intractable disease.
Conflict of interest statement
The authors declare no competing financial interests.
Figures










Comment in
-
Cancer: Double trouble for tumours.Nature. 2017 Feb 2;542(7639):34-35. doi: 10.1038/nature21117. Epub 2017 Jan 18. Nature. 2017. PMID: 28099410 No abstract available.
References
-
- Pongratz RL, Kibbey RG, Shulman GI, Cline GW. Cytosolic andmitochondrial malic enzyme isoforms differentially control insulin secretion. J Biol Chem. 2007;282:200–207. - PubMed
-
- Hutson SM, Fenstermacher D, Mahar C. Role of mitochondrial transamination in branched chain amino acid metabolism. J Biol Chem. 1988;263:3618–3625. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

