Th22 Cells Form a Distinct Th Lineage from Th17 Cells In Vitro with Unique Transcriptional Properties and Tbet-Dependent Th1 Plasticity
- PMID: 28100680
- PMCID: PMC5367520
- DOI: 10.4049/jimmunol.1601480
Th22 Cells Form a Distinct Th Lineage from Th17 Cells In Vitro with Unique Transcriptional Properties and Tbet-Dependent Th1 Plasticity
Abstract
Th22 cells are a major source of IL-22 and have been found at sites of infection and in a range of inflammatory diseases. However, their molecular characteristics and functional roles remain largely unknown because of our inability to generate and isolate pure populations. We developed a novel Th22 differentiation assay and generated dual IL-22/IL-17A reporter mice to isolate and compare pure populations of cultured Th22 and Th17 cells. Il17a fate-mapping and transcriptional profiling provide evidence that these Th22 cells have never expressed IL-17A, suggesting that they are potentially a distinct cell lineage from Th17 cells under in vitro culture conditions. Interestingly, Th22 cells also expressed granzymes, IL-13, and increased levels of Tbet. Using transcription factor-deficient cells, we demonstrate that RORγt and Tbet act as positive and negative regulators of Th22 differentiation, respectively. Furthermore, under Th1 culture conditions in vitro, as well as in an IFN-γ-rich inflammatory environment in vivo, Th22 cells displayed marked plasticity toward IFN-γ production. Th22 cells also displayed plasticity under Th2 conditions in vitro by upregulating IL-13 expression. Our work has identified conditions to generate and characterize Th22 cells in vitro. Further, it provides evidence that Th22 cells develop independently of the Th17 lineage, while demonstrating plasticity toward both Th1- and Th2-type cells.
Copyright © 2017 by The American Association of Immunologists, Inc.
Figures
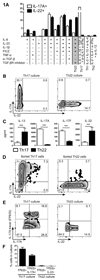
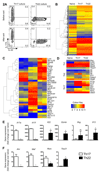
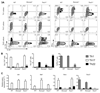


References
-
- Chung Y, Yang X, Chang SH, Ma L, Tian Q, Dong C. Expression and regulation of IL-22 in the IL-17-producing CD4+ T lymphocytes. Cell Res. 2006;16(11):902–907. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

