Insights into the RecQ helicase mechanism revealed by the structure of the helicase domain of human RECQL5
- PMID: 28100692
- PMCID: PMC5397160
- DOI: 10.1093/nar/gkw1362
Insights into the RecQ helicase mechanism revealed by the structure of the helicase domain of human RECQL5
Abstract
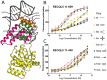
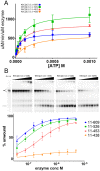
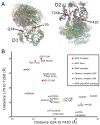
RecQ helicases are important maintainers of genome integrity with distinct roles in almost every cellular process requiring access to DNA. RECQL5 is one of five human RecQ proteins and is particularly versatile in this regard, forming protein complexes with a diverse set of cellular partners in order to coordinate its helicase activity to various processes including replication, recombination and DNA repair. In this study, we have determined crystal structures of the core helicase domain of RECQL5 both with and without the nucleotide ADP in two distinctly different ('Open' and 'Closed') conformations. Small angle X-ray scattering studies show that the 'Open' form of the protein predominates in solution and we discuss implications of this with regards to the RECQL5 mechanism and conformational changes. We have measured the ATPase, helicase and DNA binding properties of various RECQL5 constructs and variants and discuss the role of these regions and residues in the various RECQL5 activities. Finally, we have performed a systematic comparison of the RECQL5 structures with other RecQ family structures and based on these comparisons we have constructed a model for the mechano-chemical cycle of the common catalytic core of these helicases.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
-
- German J., Archibal R., Bloom D.. Chromosomal breakage in a rare and probably genetically determined syndrome of man. Science. 1965; 148:506. - PubMed
-
- Yu C.E., Oshima J., Fu Y.H., Wijsman E.M., Hisama F., Alisch R., Matthews S., Nakura J., Miki T., Ouais S. et al. . Positional cloning of the Werner's syndrome gene. Science. 1996; 272:258–262. - PubMed
-
- Kitao S., Shimamoto A., Goto M., Miller R.W., Smithson W.A., Lindor N.M., Furuichi Y.. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat. Genet. 1999; 22:82–84. - PubMed
-
- Dong Y.Z., Huang Y.X., Lu T.. Single nucleotide polymorphism in the RECQL5 gene increased osteosarcoma susceptibility in a Chinese Han population. Genet. Mol. Res. 2015; 14:1899–1902. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

