Genetic Variation in the Histamine Production, Response, and Degradation Pathway Is Associated with Histamine Pharmacodynamic Response in Children with Asthma
- PMID: 28101058
- PMCID: PMC5209333
- DOI: 10.3389/fphar.2016.00524
Genetic Variation in the Histamine Production, Response, and Degradation Pathway Is Associated with Histamine Pharmacodynamic Response in Children with Asthma
Abstract
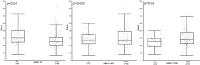
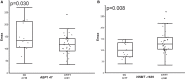
Introduction: There is growing knowledge of the wide ranging effects of histamine throughout the body therefore it is important to better understand the effects of this amine in patients with asthma. We aimed to explore the association between histamine pharmacodynamic (PD) response and genetic variation in the histamine pathway in children with asthma. Methods: Histamine Iontophoresis with Laser Doppler Monitoring (HILD) was performed in children with asthma and estimates for area under the effect curve (AUEC), maximal response over baseline (Emax), and time of Emax (Tmax) were calculated using non-compartmental analysis and non-linear mixed-effects model with a linked effect PK/PD model. DNA isolation and genotyping were performed among participants to detect known single nucleotide polymorphisms (SNPs) (n = 10) among genes (HDC, HNMT, ABP1, HRH1, HRH4) within the histamine pathway. General linear model was used to identify associations between histamine related genetic variants and measured histamine PD response parameters. Results: Genotyping and HILD response profiles were completed for 163 children. ABP1 47 C/T, ABP1 4107, and HNMT-1639 C/Twere associated with Emax (ABP1 47 CC genotype mean Emax 167.21 vs. CT/TT genotype mean Emax 139.20, p = 0.04; ABP1 4107 CC genotype mean Emax 141.72 vs. CG/GG genotype mean Emax 156.09, p = 0.005; HNMT-1639 CC genotype mean Emax 132.62 vs. CT/TT genotype mean Emax 155.3, p = 0.02). In a stratified analysis among African American children only, ABP1 and HNMT SNPs were also associated with PD response; HRH4 413 CC genotype was associated with lower Emax, p = 0.009. Conclusions: We show for the first time that histamine pathway genetic variation is associated with measureable changes in histamine response in children with asthma. The variability in histamine response and impact of histamine pathway genotype is important to further explore in patients with asthma so as to improve disease phenotyping leading to more personalized treatments.
Keywords: asthma; biomarker; histamine; pharmacodynamics; pharmacogenetics.
Figures

Similar articles
-
Genetic Variation along the Histamine Pathway in Children with Allergic versus Nonallergic Asthma.Am J Respir Cell Mol Biol. 2015 Dec;53(6):802-9. doi: 10.1165/rcmb.2014-0493OC. Am J Respir Cell Mol Biol. 2015. PMID: 25909280 Free PMC article.
-
Genetic variation within the histamine pathway among patients with asthma--a pilot study.J Asthma. 2015 May;52(4):353-62. doi: 10.3109/02770903.2014.973501. Epub 2014 Oct 30. J Asthma. 2015. PMID: 25295384 Free PMC article.
-
Polymorphisms of two histamine-metabolizing enzymes genes and childhood allergic asthma: a case control study.Clin Mol Allergy. 2010 Nov 1;8:14. doi: 10.1186/1476-7961-8-14. Clin Mol Allergy. 2010. PMID: 21040557 Free PMC article.
-
Histamine pharmacogenomics.Pharmacogenomics. 2009 May;10(5):867-83. doi: 10.2217/pgs.09.26. Pharmacogenomics. 2009. PMID: 19450133 Review.
-
Basic concepts of pharmacokinetic/pharmacodynamic (PK/PD) modelling.Int J Clin Pharmacol Ther. 1997 Oct;35(10):401-13. Int J Clin Pharmacol Ther. 1997. PMID: 9352388 Review.
Cited by
-
The impact of skin color and tone on histamine iontophoresis and Doppler flowmetry measurements as a pharmacodynamic biomarker.Clin Transl Sci. 2024 Mar;17(3):e13777. doi: 10.1111/cts.13777. Clin Transl Sci. 2024. PMID: 38511581 Free PMC article.
-
The Reliability of Histamine Pharmacodynamic Response Phenotype Classification in Children With Allergic Disease.Front Pharmacol. 2020 Mar 11;11:227. doi: 10.3389/fphar.2020.00227. eCollection 2020. Front Pharmacol. 2020. PMID: 32218731 Free PMC article.
References
-
- Akinbami L. (2006). The state of childhood asthma, United States, 1980-2005. Adv. Data 381, 1–24. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

