Importance of Radioactive Labelling to Elucidate Inositol Polyphosphate Signalling
- PMID: 28101851
- PMCID: PMC5396384
- DOI: 10.1007/s41061-016-0099-y
Importance of Radioactive Labelling to Elucidate Inositol Polyphosphate Signalling
Abstract
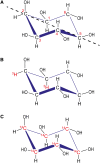
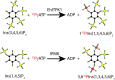
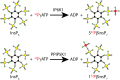
Inositol polyphosphates, in their water-soluble or lipid-bound forms, represent a large and multifaceted family of signalling molecules. Some inositol polyphosphates are well recognised as defining important signal transduction pathways, as in the case of the calcium release factor Ins(1,4,5)P3, generated by receptor activation-induced hydrolysis of the lipid PtdIns(4,5)P2 by phospholipase C. The birth of inositol polyphosphate research would not have occurred without the use of radioactive phosphate tracers that enabled the discovery of the "PI response". Radioactive labels, mainly of phosphorus but also carbon and hydrogen (tritium), have been instrumental in the development of this research field and the establishment of the inositol polyphosphates as one of the most important networks of regulatory molecules present in eukaryotic cells. Advancements in microscopy and mass spectrometry and the development of colorimetric assays have facilitated inositol polyphosphate research, but have not eliminated the need for radioactive experimental approaches. In fact, such experiments have become easier with the cloning of the inositol polyphosphate kinases, enabling the systematic labelling of specific positions of the inositol ring with radioactive phosphate. This approach has been valuable for elucidating their metabolic pathways and identifying specific and novel functions for inositol polyphosphates. For example, the synthesis of radiolabelled inositol pyrophosphates has allowed the discovery of a new protein post-translational modification. Therefore, radioactive tracers have played and will continue to play an important role in dissecting the many complex aspects of inositol polyphosphate physiology. In this review we aim to highlight the historical importance of radioactivity in inositol polyphosphate research, as well as its modern usage.
Keywords: Inositol; Metabolism; Phosphate; Pyrophosphates; Radioactivity.
Figures


Similar articles
-
[3H]inositol polyphosphate metabolism in muscarinic cholinoceptor-stimulated airways smooth muscle: accumulation of [3H]inositol 4,5 bisphosphate via a lithium-sensitive inositol polyphosphate 1-phosphatase.J Pharmacol Exp Ther. 1997 Feb;280(2):974-82. J Pharmacol Exp Ther. 1997. PMID: 9023314
-
Phosphate, inositol and polyphosphates.Biochem Soc Trans. 2016 Feb;44(1):253-9. doi: 10.1042/BST20150215. Biochem Soc Trans. 2016. PMID: 26862212 Review.
-
Are inositol pyrophosphates signalling molecules?J Cell Physiol. 2009 Jul;220(1):8-15. doi: 10.1002/jcp.21763. J Cell Physiol. 2009. PMID: 19326391 Review.
-
Inositol polyphosphate metabolism and inositol lipids in a green alga, Chlamydomonas eugametos.Biochem J. 1992 Jan 1;281 ( Pt 1)(Pt 1):261-6. doi: 10.1042/bj2810261. Biochem J. 1992. PMID: 1310008 Free PMC article.
-
The inositol pyrophosphate synthesis pathway in Trypanosoma brucei is linked to polyphosphate synthesis in acidocalcisomes.Mol Microbiol. 2017 Oct;106(2):319-333. doi: 10.1111/mmi.13766. Epub 2017 Aug 22. Mol Microbiol. 2017. PMID: 28792096 Free PMC article.
Cited by
-
The phytase RipBL1 enables the assignment of a specific inositol phosphate isomer as a structural component of human kidney stones.RSC Chem Biol. 2023 Jan 27;4(4):300-309. doi: 10.1039/d2cb00235c. eCollection 2023 Apr 5. RSC Chem Biol. 2023. PMID: 37034402 Free PMC article.
-
The poly(A) polymerase Star-PAP is regulated by stably associated phosphoinositide messengers.J Biol Chem. 2025 Jun 24;301(8):110412. doi: 10.1016/j.jbc.2025.110412. Online ahead of print. J Biol Chem. 2025. PMID: 40570960 Free PMC article.
-
Pools of Independently Cycling Inositol Phosphates Revealed by Pulse Labeling with 18O-Water.J Am Chem Soc. 2025 May 28;147(21):17626-17641. doi: 10.1021/jacs.4c16206. Epub 2025 May 15. J Am Chem Soc. 2025. PMID: 40372010 Free PMC article.
-
Myo-Inositol and D-Chiro-Inositol as Modulators of Ovary Steroidogenesis: A Narrative Review.Nutrients. 2023 Apr 13;15(8):1875. doi: 10.3390/nu15081875. Nutrients. 2023. PMID: 37111094 Free PMC article. Review.
-
Analysis of inositol phosphate metabolism by capillary electrophoresis electrospray ionization mass spectrometry.Nat Commun. 2020 Nov 27;11(1):6035. doi: 10.1038/s41467-020-19928-x. Nat Commun. 2020. PMID: 33247133 Free PMC article.
References
-
- Menniti FS, et al. Turnover of inositol polyphosphate pyrophosphates in pancreatoma cells. J Biol Chem. 1993;268(6):3850–3856. - PubMed
-
- Stephens L, et al. The detection, purification, structural characterization, and metabolism of diphosphoinositol pentakisphosphate(s) and bisdiphosphoinositol tetrakisphosphate(s) J Biol Chem. 1993;268(6):4009–4015. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
