Chelerythrine down regulates expression of VEGFA, BCL2 and KRAS by arresting G-Quadruplex structures at their promoter regions
- PMID: 28102286
- PMCID: PMC5244364
- DOI: 10.1038/srep40706
Chelerythrine down regulates expression of VEGFA, BCL2 and KRAS by arresting G-Quadruplex structures at their promoter regions
Abstract
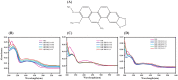
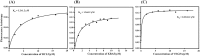
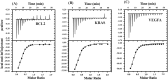
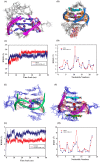
A putative anticancer plant alkaloid, Chelerythrine binds to G-quadruplexes at promoters of VEGFA, BCL2 and KRAS genes and down regulates their expression. The association of Chelerythrine to G-quadruplex at the promoters of these oncogenes were monitored using UV absorption spectroscopy, fluorescence anisotropy, circular dichroism spectroscopy, CD melting, isothermal titration calorimetry, molecular dynamics simulation and quantitative RT-PCR technique. The pronounced hypochromism accompanied by red shifts in UV absorption spectroscopy in conjunction with ethidium bromide displacement assay indicates end stacking mode of interaction of Chelerythrine with the corresponding G-quadruplex structures. An increase in fluorescence anisotropy and CD melting temperature of Chelerythrine-quadruplex complex revealed the formation of stable Chelerythrine-quadruplex complex. Isothermal titration calorimetry data confirmed that Chelerythrine-quadruplex complex formation is thermodynamically favourable. Results of quantative RT-PCR experiment in combination with luciferase assay showed that Chelerythrine treatment to MCF7 breast cancer cells effectively down regulated transcript level of all three genes, suggesting that Chelerythrine efficiently binds to in cellulo quadruplex motifs. MD simulation provides the molecular picture showing interaction between Chelerythrine and G-quadruplex. Binding of Chelerythrine with BCL2, VEGFA and KRAS genes involved in evasion, angiogenesis and self sufficiency of cancer cells provides a new insight for the development of future therapeutics against cancer.
Figures




References
-
- Watson J. D. & Crick F. H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 171, 737–738 (1953). - PubMed
-
- Brooks T. A. & Hurley L. H. The role of supercoiling in transcriptional control of MYC and its importance in molecular therapeutics. Nat Rev Cancer 9, 849–861 (2009). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

