Genome engineering of stem cell organoids for disease modeling
- PMID: 28102490
- PMCID: PMC5413597
- DOI: 10.1007/s13238-016-0368-0
Genome engineering of stem cell organoids for disease modeling
Abstract
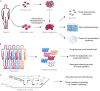
Precision medicine emerges as a new approach that takes into account individual variability. Successful realization of precision medicine requires disease models that are able to incorporate personalized disease information and recapitulate disease development processes at the molecular, cellular and organ levels. With recent development in stem cell field, a variety of tissue organoids can be derived from patient specific pluripotent stem cells and adult stem cells. In combination with the state-of-the-art genome editing tools, organoids can be further engineered to mimic disease-relevant genetic and epigenetic status of a patient. This has therefore enabled a rapid expansion of sophisticated in vitro disease models, offering a unique system for fundamental and biomedical research as well as the development of personalized medicine. Here we summarize some of the latest advances and future perspectives in engineering stem cell organoids for human disease modeling.
Keywords: genome editing; pluripotent/adult stem cell; precision medicine; tissue organoid.
Figures

References
-
- Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, van Es JH, Sato T, Stange DE, Begthel H, van den Born M, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. - PubMed
-
- Ben-Zvi D, Melton DA. Modeling human nutrition using human embryonic stem cells. Cell. 2015;161:12–17. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

