Structural Analysis of the Glycosylated Intact HIV-1 gp120-b12 Antibody Complex Using Hydroxyl Radical Protein Footprinting
- PMID: 28102671
- PMCID: PMC5319886
- DOI: 10.1021/acs.biochem.6b00888
Structural Analysis of the Glycosylated Intact HIV-1 gp120-b12 Antibody Complex Using Hydroxyl Radical Protein Footprinting
Abstract
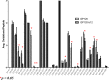
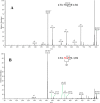
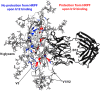
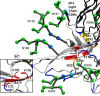
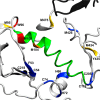
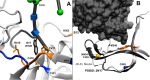
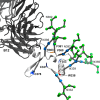
Glycoprotein gp120 is a surface antigen and virulence factor of human immunodeficiency virus 1. Broadly neutralizing antibodies (bNAbs) that react to gp120 from a variety of HIV isolates offer hope for the development of broadly effective immunogens for vaccination purposes, if the interactions between gp120 and bNAbs can be understood. From a structural perspective, gp120 is a particularly difficult system because of its size, the presence of multiple flexible regions, and the large amount of glycosylation, all of which are important in gp120-bNAb interactions. Here, the interaction of full-length, glycosylated gp120 with bNAb b12 is probed using high-resolution hydroxyl radical protein footprinting (HR-HRPF) by fast photochemical oxidation of proteins. HR-HRPF allows for the measurement of changes in the average solvent accessible surface area of multiple amino acids without the need for measures that might alter the protein conformation, such as mutagenesis. HR-HRPF of the gp120-b12 complex coupled with computational modeling shows a novel extensive interaction of the V1/V2 domain, probably with the light chain of b12. Our data also reveal HR-HRPF protection in the C3 domain caused by interaction of the N330 glycan with the b12 light chain. In addition to providing information about the interactions of full-length, glycosylated gp120 with b12, this work serves as a template for the structural interrogation of full-length glycosylated gp120 with other bNAbs to better characterize the interactions that drive the broad specificity of the bNAb.
Conflict of interest statement
The authors declare the following competing financial interest(s): J.S.S. discloses a significant ownership share of Photochem Technologies, LLC, a small company active in the area of hydroxyl radical protein footprinting.
Figures

References
-
- Nara P. L.; Garrity R. R.; Goudsmit J. (1991) Neutralization of HIV-1: a paradox of humoral proportions. FASEB J. 5, 2437–2455. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

