Basis of catalytic assembly of the mitotic checkpoint complex
- PMID: 28102834
- PMCID: PMC5448665
- DOI: 10.1038/nature21384
Basis of catalytic assembly of the mitotic checkpoint complex
Abstract
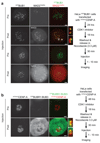
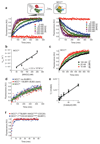
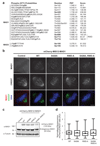
In mitosis, for each daughter cell to inherit an accurate copy of the genome from the mother cell, sister chromatids in the mother cell must attach to microtubules emanating from opposite poles of the mitotic spindle, a process known as bi-orientation. A surveillance mechanism, termed the spindle assembly checkpoint (SAC), monitors the microtubule attachment process and can temporarily halt the separation of sister chromatids and the completion of mitosis until bi-orientation is complete. SAC failure results in abnormal chromosome numbers, termed aneuploidy, in the daughter cells, a hallmark of many tumours. The HORMA-domain-containing protein mitotic arrest deficient 2 (MAD2) is a subunit of the SAC effector mitotic checkpoint complex (MCC). Structural conversion from the open to the closed conformation of MAD2 is required for MAD2 to be incorporated into the MCC. In vitro, MAD2 conversion and MCC assembly take several hours, but in cells the SAC response is established in a few minutes. Here, to address this discrepancy, we reconstituted a near-complete SAC signalling system with purified components and monitored assembly of the MCC in real time. A marked acceleration in MAD2 conversion and MCC assembly was observed when monopolar spindle 1 (MPS1) kinase phosphorylated the MAD1-MAD2 complex, triggering it to act as the template for MAD2 conversion and therefore contributing to the establishment of a physical platform for MCC assembly. Thus, catalytic activation of the SAC depends on regulated protein-protein interactions that accelerate the spontaneous but rate-limiting conversion of MAD2 required for MCC assembly.
Figures









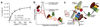
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

