Protein Phosphatase 1 Down Regulates ZYG-1 Levels to Limit Centriole Duplication
- PMID: 28103229
- PMCID: PMC5289615
- DOI: 10.1371/journal.pgen.1006543
Protein Phosphatase 1 Down Regulates ZYG-1 Levels to Limit Centriole Duplication
Abstract
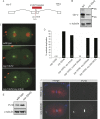
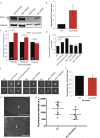
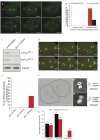
In humans perturbations of centriole number are associated with tumorigenesis and microcephaly, therefore appropriate regulation of centriole duplication is critical. The C. elegans homolog of Plk4, ZYG-1, is required for centriole duplication, but our understanding of how ZYG-1 levels are regulated remains incomplete. We have identified the two PP1 orthologs, GSP-1 and GSP-2, and their regulators I-2SZY-2 and SDS-22 as key regulators of ZYG-1 protein levels. We find that down-regulation of PP1 activity either directly, or by mutation of szy-2 or sds-22 can rescue the loss of centriole duplication associated with a zyg-1 hypomorphic allele. Suppression is achieved through an increase in ZYG-1 levels, and our data indicate that PP1 normally regulates ZYG-1 through a post-translational mechanism. While moderate inhibition of PP1 activity can restore centriole duplication to a zyg-1 mutant, strong inhibition of PP1 in a wild-type background leads to centriole amplification via the production of more than one daughter centriole. Our results thus define a new pathway that limits the number of daughter centrioles produced each cycle.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
The E2F-DP1 Transcription Factor Complex Regulates Centriole Duplication in Caenorhabditis elegans.G3 (Bethesda). 2016 Jan 15;6(3):709-20. doi: 10.1534/g3.115.025577. G3 (Bethesda). 2016. PMID: 26772748 Free PMC article.
-
Control of mitotic and meiotic centriole duplication by the Plk4-related kinase ZYG-1.J Cell Sci. 2010 Mar 1;123(Pt 5):795-805. doi: 10.1242/jcs.050682. Epub 2010 Feb 9. J Cell Sci. 2010. PMID: 20144993 Free PMC article.
-
The chromatin remodeling protein CHD-1 and the EFL-1/DPL-1 transcription factor cooperatively down regulate CDK-2 to control SAS-6 levels and centriole number.PLoS Genet. 2022 Apr 4;18(4):e1009799. doi: 10.1371/journal.pgen.1009799. eCollection 2022 Apr. PLoS Genet. 2022. PMID: 35377871 Free PMC article.
-
Centrosome duplication and nematodes: recent insights from an old relationship.Dev Cell. 2005 Sep;9(3):317-25. doi: 10.1016/j.devcel.2005.08.004. Dev Cell. 2005. PMID: 16139223 Review.
-
Finding treasures in frozen cells: new centriole intermediates.Bioessays. 2007 Jul;29(7):630-4. doi: 10.1002/bies.20594. Bioessays. 2007. PMID: 17563074 Review.
Cited by
-
Genetic Modifiers and Rare Mendelian Disease.Genes (Basel). 2020 Feb 25;11(3):239. doi: 10.3390/genes11030239. Genes (Basel). 2020. PMID: 32106447 Free PMC article. Review.
-
A membrane reticulum, the centriculum, affects centrosome size and function in Caenorhabditis elegans.Curr Biol. 2023 Mar 13;33(5):791-806.e7. doi: 10.1016/j.cub.2022.12.059. Epub 2023 Jan 23. Curr Biol. 2023. PMID: 36693370 Free PMC article.
-
Site-specific phosphorylation of ZYG-1 regulates ZYG-1 stability and centrosome number.iScience. 2023 Nov 8;26(12):108410. doi: 10.1016/j.isci.2023.108410. eCollection 2023 Dec 15. iScience. 2023. PMID: 38034351 Free PMC article.
-
APC/CFZR-1 regulates centrosomal ZYG-1 to limit centrosome number.J Cell Sci. 2021 Jul 15;134(14):jcs253088. doi: 10.1242/jcs.253088. Epub 2021 Jul 26. J Cell Sci. 2021. PMID: 34308970 Free PMC article.
-
Protease dead separase inhibits chromosome segregation and RAB-11 vesicle trafficking.Cell Cycle. 2017 Oct 18;16(20):1902-1917. doi: 10.1080/15384101.2017.1363936. Epub 2017 Aug 18. Cell Cycle. 2017. PMID: 28820333 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

