miRNAs Do Not Regulate Circadian Protein Synthesis in the Dinoflagellate Lingulodinium polyedrum
- PMID: 28103286
- PMCID: PMC5245829
- DOI: 10.1371/journal.pone.0168817
miRNAs Do Not Regulate Circadian Protein Synthesis in the Dinoflagellate Lingulodinium polyedrum
Abstract
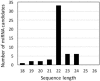
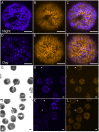
Dinoflagellates have been shown to express miRNA by bioinformatics and RNA blot (Northern) analyses. However, it is not yet known if miRNAs are able to alter gene expression in this class of organisms. We have assessed the possibility that miRNA may mediate circadian regulation of gene expression in the dinoflagellate Lingulodinium polyedrum using the Luciferin Binding Protein (LBP) as a specific example. LBP is a good candidate for regulation by miRNA since mRNA levels are constant over the daily cycle while protein synthesis is restricted by the circadian clock to a period of several hours at the start of the night phase. The transcriptome contains a potential DICER and an ARGONAUTE, suggesting the machinery for generating miRNAs is present. Furthermore, a probe directed against an abundant Symbiodinium miRNA cross reacts on Northern blots. However, L. polyedrum has no small RNAs detectable by ethidium bromide staining, even though higher plant miRNAs run in parallel are readily observed. Illumina sequencing of small RNAs showed that the majority of reads did not have a match in the L. polyedrum transcriptome, and those that did were almost all sense strand mRNA fragments. A direct search for 18-26 nucleotide long RNAs capable of forming duplexes with a 2 base 3' overhang detected 53 different potential miRNAs, none of which was able to target any of the known circadian regulated genes. Lastly, a microscopy-based test to assess synthesis of the naturally fluorescent LBP in single cells showed that neither double-stranded nor antisense lbp RNA introduced into cells by microparticle bombardment prior to the time of LBP synthesis were able to reduce the amount of LBP produced. Taken together, our results indicate that circadian control of protein synthesis in L. polyedrum is not mediated by miRNAs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Nassoury N, Hastings JW, Morse D. Biochemistry and circadian regulation of output from the Gonyaulax clock: are there many clocks or simply many hands? In: Kippert F, editor. The circadian clock in eukaryotic microbes: Kluwer Academic and Plenum; 2005.
-
- Nicolas MT, Morse D, Bassot JM, Hastings JW. Colocalization of luciferin binding protein and luciferase to the scintillons of Gonyaulax polyedra revealed by double immunolabeling after fast freeze fixation. Protoplasma. 1991;160:159–66.
-
- Fritz L, Morse D, Hastings JW. The circadian bioluminescence rhythm of Gonyaulax is related to daily variations in the number of light-emitting organelles. J Cell Sci. 1990;95(Pt 2):321–8. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

