Host-microbiota interactions: epigenomic regulation
- PMID: 28103497
- PMCID: PMC5451311
- DOI: 10.1016/j.coi.2016.12.001
Host-microbiota interactions: epigenomic regulation
Abstract
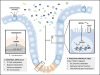
The coevolution of mammalian hosts and their commensal microbiota has led to the development of complex symbiotic relationships between resident microbes and mammalian cells. Epigenomic modifications enable host cells to alter gene expression without modifying the genetic code, and therefore represent potent mechanisms by which mammalian cells can transcriptionally respond, transiently or stably, to environmental cues. Advances in genome-wide approaches are accelerating our appreciation of microbial influences on host physiology, and increasing evidence highlights that epigenomics represent a level of regulation by which the host integrates and responds to microbial signals. In particular, bacterial-derived short chain fatty acids have emerged as one clear link between how the microbiota intersects with host epigenomic pathways. Here we review recent findings describing crosstalk between the microbiota and epigenomic pathways in multiple mammalian cell populations. Further, we discuss interesting links that suggest that the scope of our understanding of epigenomic regulation in the host-microbiota relationship is still in its infancy.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

