Immune Receptors and Co-receptors in Antiviral Innate Immunity in Plants
- PMID: 28105028
- PMCID: PMC5214455
- DOI: 10.3389/fmicb.2016.02139
Immune Receptors and Co-receptors in Antiviral Innate Immunity in Plants
Abstract
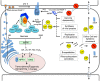
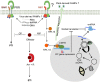
Plants respond to pathogens using an innate immune system that is broadly divided into PTI (pathogen-associated molecular pattern- or PAMP-triggered immunity) and ETI (effector-triggered immunity). PTI is activated upon perception of PAMPs, conserved motifs derived from pathogens, by surface membrane-anchored pattern recognition receptors (PRRs). To overcome this first line of defense, pathogens release into plant cells effectors that inhibit PTI and activate effector-triggered susceptibility (ETS). Counteracting this virulence strategy, plant cells synthesize intracellular resistance (R) proteins, which specifically recognize pathogen effectors or avirulence (Avr) factors and activate ETI. These coevolving pathogen virulence strategies and plant resistance mechanisms illustrate evolutionary arms race between pathogen and host, which is integrated into the zigzag model of plant innate immunity. Although antiviral immune concepts have been initially excluded from the zigzag model, recent studies have provided several lines of evidence substantiating the notion that plants deploy the innate immune system to fight viruses in a manner similar to that used for non-viral pathogens. First, most R proteins against viruses so far characterized share structural similarity with antibacterial and antifungal R gene products and elicit typical ETI-based immune responses. Second, virus-derived PAMPs may activate PTI-like responses through immune co-receptors of plant PTI. Finally, and even more compelling, a viral Avr factor that triggers ETI in resistant genotypes has recently been shown to act as a suppressor of PTI, integrating plant viruses into the co-evolutionary model of host-pathogen interactions, the zigzag model. In this review, we summarize these important progresses, focusing on the potential significance of antiviral immune receptors and co-receptors in plant antiviral innate immunity. In light of the innate immune system, we also discuss a newly uncovered layer of antiviral defense that is specific to plant DNA viruses and relies on transmembrane receptor-mediated translational suppression for defense.
Keywords: ETI; NSP-Interacting kinase 1; PAMP-triggered immunity; PTI; antiviral immunity; effector-triggered immunity; receptor NIK1; resistance genes.
Figures
References
-
- Aarts N., Metz M., Holub E., Staskawicz B. J., Daniels M. J., Parker J. E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 95 10306–10311. 10.1073/pnas.95.17.10306 - DOI - PMC - PubMed
-
- Anagnostou K., Jahn M., Perl-Treves R. (2000). Inheritance and linkage analysis of resistance to Zucchini yellow mosaic virus, Watermelon mosaic virus, Papaya ringspot virus and powdery mildew in melon. Euphytica 116 265–270. 10.1023/A:1004005716806 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

