A Grapevine TTG2-Like WRKY Transcription Factor Is Involved in Regulating Vacuolar Transport and Flavonoid Biosynthesis
- PMID: 28105033
- PMCID: PMC5214514
- DOI: 10.3389/fpls.2016.01979
A Grapevine TTG2-Like WRKY Transcription Factor Is Involved in Regulating Vacuolar Transport and Flavonoid Biosynthesis
Abstract
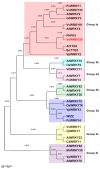
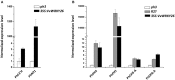
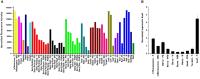
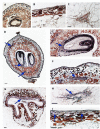
A small set of TTG2-like homolog proteins from different species belonging to the WRKY family of transcription factors were shown to share a similar mechanism of action and to control partially conserved biochemical/developmental processes in their native species. In particular, by activating P-ATPases residing on the tonoplast, PH3 from Petunia hybrida promotes vacuolar acidification in petal epidermal cells whereas TTG2 from Arabidopsis thaliana enables the accumulation of proanthocyanidins in the seed coat. In this work we functionally characterized VvWRKY26 identified as the closest grapevine homolog of PhPH3 and AtTTG2. When constitutively expressed in petunia ph3 mutant, VvWRKY26 can fulfill the PH3 function in the regulation of vacuolar pH and restores the wild type pigmentation phenotype. By a global correlation analysis of gene expression and by transient over-expression in Vitis vinifera, we showed transcriptomic relationships of VvWRKY26 with many genes related to vacuolar acidification and transport in grapevine. Moreover, our results indicate an involvement in flavonoid pathway possibly restricted to the control of proanthocyanidin biosynthesis that is consistent with its expression pattern in grape berry tissues. Overall, the results show that, in addition to regulative mechanisms and biological roles shared with TTG2-like orthologs, VvWRKY26 can play roles in fleshy fruit development that have not been previously reported in studies from dry fruit species. This study paves the way toward the comprehension of the regulatory network controlling vacuolar acidification and flavonoid accumulation mechanisms that contribute to the final berry quality traits in grapevine.
Keywords: WRKY; flavonoids; grapevine; petunia; vacuolar acidification.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

