Loss of the SWI/SNF ATPase subunits BRM and BRG1 drives lung cancer development
- PMID: 28105457
- PMCID: PMC5235921
- DOI: 10.18632/oncoscience.323
Loss of the SWI/SNF ATPase subunits BRM and BRG1 drives lung cancer development
Abstract
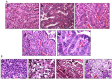
Inactivation of Brg1 and Brm accelerated lung tumor development, shortened tumor latency, and caused a loss of differentiation. Tumors with Brg1 and/or Brm loss recapitulated the evolution of human lung cancer as observed by the development of local tumor invasion as well as distal tumor metastasis, thereby making this model useful in lung cancer studies. Brg1 loss contributed to metastasis in part by driving E-cadherin loss and Vimentin up-regulation. By changing more than 6% of the murine genome with the down-regulation of tumor suppressors, DNA repair, differentiation and cell adhesion genes, and the concomitant up-regulation of oncogenes, angiogenesis, metastasis and antiapoptosis genes, caused by the dual loss of Brg1/Brm further accelerated tumor development. Additionally, this Brg1/Brm-driven change in gene expression resulted in a nearly two-fold increase in tumorigenicity in Brg1/Brm knockout mice compared with wild type mice. Most importantly, Brg1/Brm-driven lung cancer development histologically and clinically reflects human lung cancer development thereby making this GEMM model potentially useful.
Keywords: lung cancer; smarca2; smarca4; swi/snf; tumor.
Conflict of interest statement
There are no conflicts of interest to disclose. Dr Reisman is the CEO of Zenegene Inc.
Figures






References
-
- Peterson CL, Workman JL. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev. 2000;10:187–192. - PubMed
-
- Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol. 1984;178:853–868. - PubMed
-
- Peterson CL, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. - PubMed
-
- Yoshinaga SK, Peterson CL, Herskowitz I, Yamamoto KR. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992;258:1598–1604. - PubMed
-
- Carlson M, Laurent BC. The SNF/SWI family of global transcriptional activators. Curr Opin Cell Biol. 1994;6:396–402. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

