Genes of susceptibility to early neurodegenerative changes in the rat retina and brain: analysis by means of congenic strains
- PMID: 28105932
- PMCID: PMC5249004
- DOI: 10.1186/s12863-016-0461-7
Genes of susceptibility to early neurodegenerative changes in the rat retina and brain: analysis by means of congenic strains
Abstract
Background: There has been considerable interest in discovery of the genetic architecture of complex traits, particularly age-related neurodegenerative disorders. To predict disease risk and to understand its genetic basis in humans, it is necessary to study animal models. Our previous research on the accelerated-senescence OXYS strain has revealed two quantitative trait loci (QTLs) on rat chromosome 1 that are associated with early cataract and/or retinopathy as well as with behavioral abnormalities. Each locus was partially mapped within the introgressed segments in a certain congenic strain: WAG/OXYS-1.1 or WAG/OXYS-1.2. Retinal transcriptome profiling of 20-day-old congenic and OXYS rats by high-throughput RNA sequencing uncovered relevant candidate genes and pathways. Nonetheless, the question remained open whether the same genetic components simultaneously have effects on various manifestations of the accelerated-senescence phenotype in OXYS rats. The present study was designed to analyze the genes of susceptibility to early neurodegenerative processes taking place in the OXYS rat retina and brain and to assess their potential functional clustering. The study was based on the findings from recent publications (including mapping of quantitative trait loci) and on comparative phenotyping of congenic rat strains.
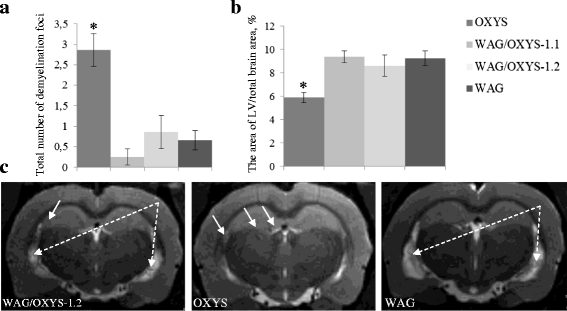
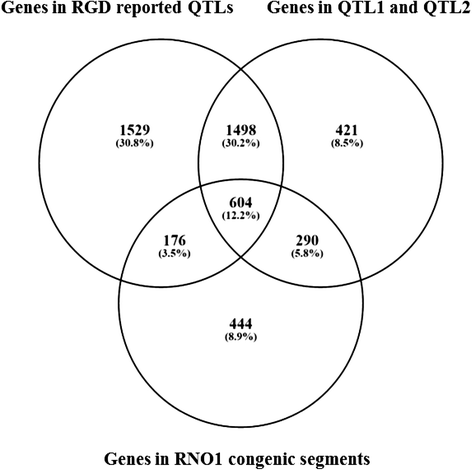
Results: The backcrossing of Wistar Albino Glaxo (WAG) and OXYS strains to generate the congenics resulted in two congenic strains with high susceptibility to cataract and retinopathy but with no obvious signs of Alzheimer's disease-like brain pathology that are specific for OXYS rats. Thus, the genes of susceptibility to brain neurodegeneration were not introgressed into the congenic strains or there is a strong effect of the genetic background on the disease phenotype. Moreover, the progression of retinopathy with age was relatively less severe in the WAG background compared to the OXYS background. A comparative analysis of previously defined QTLs and congenic segments led to identification of candidate genes with a suspected effect on brain neurodegeneration including the genes showing differential expression in the congenic strains.
Conclusion: Overall, our findings suggest that the cause of the cataract and the cause of retinopathy phenotypes in OXYS rats may be genetically linked to each other within the introgressed segments in the WAG/OXYS-1.1 and/or WAG/OXYS-1.2 congenic strains.
Keywords: Age-related macular degeneration; Alzheimer’s disease; Congenic strain; Genetic architecture of complex trait; OXYS rats; Quantitative trait locus.
Figures



References
-
- Ackert-Bicknell CL, Anderson LC, Sheehan S, Hill WG, Chang B, Churchill GA, et al. Aging Research Using Mouse Models. Curr. Protoc. Mouse Biol. [Internet]. Hoboken, NJ, USA: John Wiley & Sons, Inc.; 2015. p. 95–133. Available from: http://doi.wiley.com/10.1002/9780470942390.mo140195. - PMC - PubMed
-
- Jellinger KA. Basic mechanisms of neurodegeneration: a critical update. J. Cell. Mol. Med. [Internet]. 2010; Available from: http://doi.wiley.com/10.1111/j.1582-4934.2010.01010.x. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

