The occurrence of spring forms in tetraploid Timopheevi wheat is associated with variation in the first intron of the VRN-A1 gene
- PMID: 28105942
- PMCID: PMC5123382
- DOI: 10.1186/s12870-016-0925-y
The occurrence of spring forms in tetraploid Timopheevi wheat is associated with variation in the first intron of the VRN-A1 gene
Abstract
Background: Triticum araraticum and Triticum timopheevii are tetraploid species of the Timopheevi group. The former includes both winter and spring forms with a predominance of winter forms, whereas T. timopheevii is considered a spring species. In order to clarify the origin of the spring growth habit in T. timopheevii, allelic variability of the VRN-1 gene was investigated in a set of accessions of both tetraploid species, together with the diploid species Ae. speltoides, presumed donor of the G genome to these tetraploids.
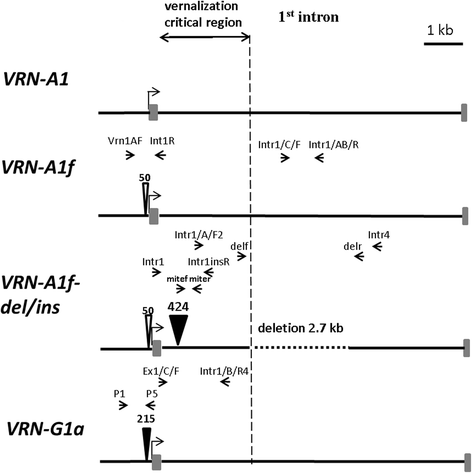
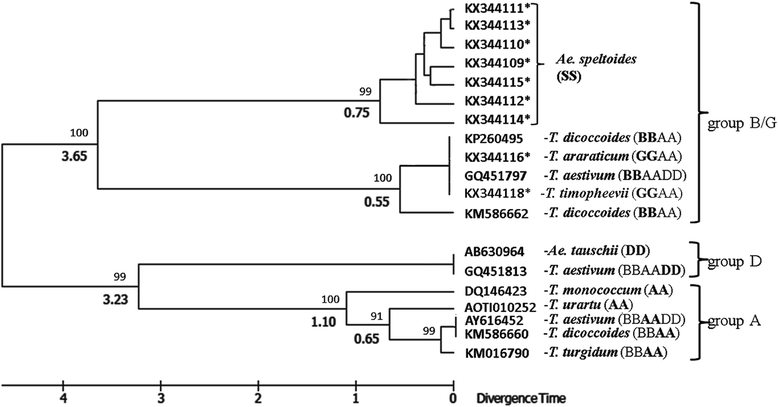
Results: The promoter region of the VRN-A1 locus in all studied tetraploid accessions of both T. araraticum and T. timopheevii represents the previously described allele VRN-A1f with a 50 bp deletion near the start codon. Three additional alleles were identified namely, VRN-A1f-del, VRN-A1f-ins and VRN-A1f-del/ins, which contained large mutations in the first (1st) intron of VRN-A1. The first allele, carrying a deletion of 2.7 kb in a central part of intron 1, occurred in a few accessions of T. araraticum and no accessions of T. timopheevii. The VRN-A1f-ins allele, containing the insertion of a 0.4 kb MITE element about 0.4 kb upstream from the start of intron 1, and allele VRN-A1f-del/ins having this insertion coupled with a deletion of 2.7 kb are characteristic only for T. timopheevii. Allelic variation at the VRN-G1 locus includes the previously described allele VRN-G1a (with the insertion of a 0.2 kb MITE in the promoter) found in a few accessions of both tetraploid species. We showed that alleles VRN-A1f-del and VRN-G1a have no association with the spring growth habit, while in all accessions of T. timopheevii this habit was associated with the dominant VRN-A1f-ins and VRN-A1f-del/ins alleles. None of the Ae. speltoides accessions included in this study had changes in the promoter or 1st intron regions of VRN-1 which might confer a spring growth habit. The VRN-1 promoter sequences analyzed herein and downloaded from databases have been used to construct a phylogram to assess the time of divergence of Ae. speltoides in relation to other wheat species.
Conclusions: Among accessions of T. araraticum, the preferentially winter predecessor of T. timopheevii, two large mutations were found in both VRN-A1 and VRN-G1 loci (VRN-A1f-del and VRN-G1a) that were found to have no effect on vernalization requirements. Spring tetraploid T. timopheevii had one VRN-1 allele in common for two species (VRN-G1a), and two that were specific (VRN-A1f-ins, VRN-A1f-del/ins). The latter alleles include mutations in the 1st intron of VRN-A1 and also share a 0.4 kb MITE insertion near the start of intron 1. We suggested that this insertion resulted in a spring growth habit in a progenitor of T. timopheevii which has probably been selected during subsequent domestication. The phylogram constructed on the basis of the VRN-1 promoter sequences confirmed the early divergence (~3.5 MYA) of the ancestor(s) of the B/G genomes from Ae. speltoides.
Keywords: Aegilops; Allelic variation; First intron; Promoter; Triticum; VRN-1 gene; Vernalization.
Figures


References
-
- Huang S, Sirikhachornkit A, Su X, Faris J, Gill B, Haselkorn R, Gornicki P. Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc Natl Acad Sci U S A. 2002;99:8133–8138. doi: 10.1073/pnas.072223799. - DOI - PMC - PubMed
-
- Badaeva ED, Boguslavsky RL, Badaev NS, Zelenin AV. Intraspecific chromosomal polymorphism of Triticum araraticum (Poaceae) detected by C-banding technique. P1 Syst Evol. 1990;169:13–24. doi: 10.1007/BF00935980. - DOI
-
- Goncharov NP. Comparative genetics of wheats and their related species. Novosibirsk: Siberian University Press (In Russian); 2002. p. 251.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

