VRN1 genes variability in tetraploid wheat species with a spring growth habit
- PMID: 28105956
- PMCID: PMC5123248
- DOI: 10.1186/s12870-016-0924-z
VRN1 genes variability in tetraploid wheat species with a spring growth habit
Abstract
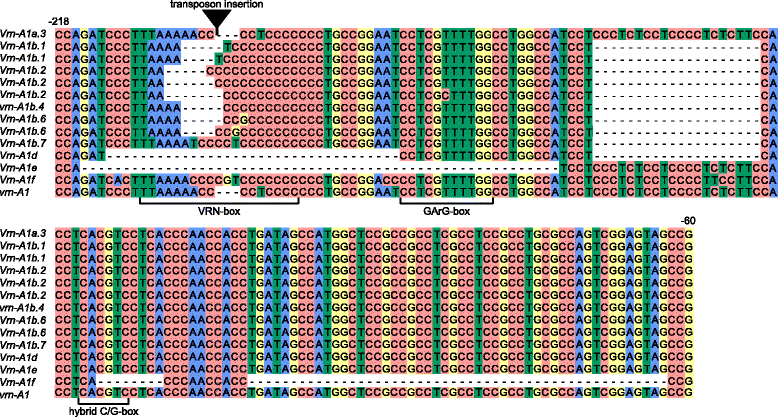
Background: Vernalization genes VRN1 play a major role in the transition from vegetative to reproductive growth in wheat. In di-, tetra- and hexaploid wheats the presence of a dominant allele of at least one VRN1 gene homologue (Vrn-A1, Vrn-B1, Vrn-G1 or Vrn-D1) determines the spring growth habit. Allelic variation between the Vrn-1 and vrn-1 alleles relies on mutations in the promoter region or the first intron. The origin and variability of the dominant VRN1 alleles, determining the spring growth habit in tetraploid wheat species have been poorly studied.
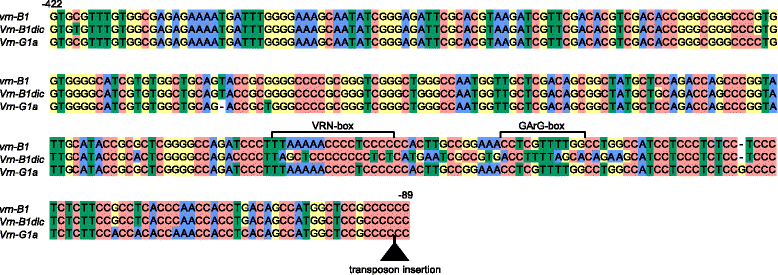
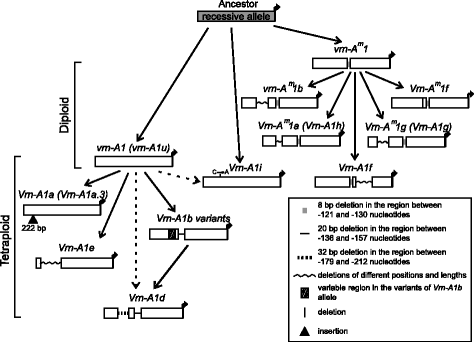
Results: Here we analyzed the growth habit of 228 tetraploid wheat species accessions and 25 % of them were spring type. We analyzed the promoter and first intron regions of VRN1 genes in 57 spring accessions of tetraploid wheats. The spring growth habit of most studied spring accessions was determined by previously identified dominant alleles of VRN1 genes. Genetic experiments proof the dominant inheritance of Vrn-A1d allele which was widely distributed across the accessions of Triticum dicoccoides. Two novel alleles were discovered and designated as Vrn-A1b.7 and Vrn-B1dic. Vrn-A1b.7 had deletions of 20 bp located 137 bp upstream of the start codon and mutations within the VRN-box when compared to the recessive allele of vrn-A1. So far the Vrn-A1d allele was identified only in spring accessions of the T. dicoccoides and T. turgidum species. Vrn-B1dic was identified in T. dicoccoides IG46225 and had 11 % sequence dissimilarity in comparison to the promoter of vrn-B1. The presence of Vrn-A1b.7 and Vrn-B1dic alleles is a predicted cause of the spring growth habit of studied accessions of tetraploid species. Three spring accessions T. aethiopicum K-19059, T. turanicum K-31693 and T. turgidum cv. Blancal possess recessive alleles of both VRN-A1 and VRN-B1 genes. Further investigations are required to determine the source of spring growth habit of these accessions.
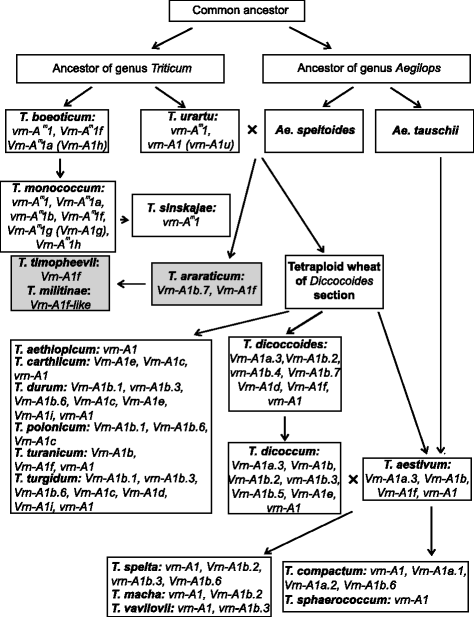
Conclusions: New allelic variants of the VRN-A1 and VRN-B1 genes were identified in spring accessions of tetraploid wheats. The origin and evolution of VRN-A1 alleles in di- and tetraploid wheat species was discussed.
Keywords: Evolution; Growth habit; Triticum; VRN1 gene; Wheat; vernalization.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

