Identification of nuclear genes controlling chlorophyll synthesis in barley by RNA-seq
- PMID: 28105957
- PMCID: PMC5123340
- DOI: 10.1186/s12870-016-0926-x
Identification of nuclear genes controlling chlorophyll synthesis in barley by RNA-seq
Abstract
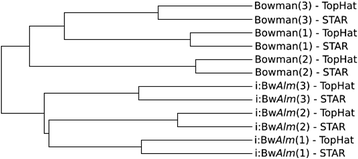
Background: Albinism in plants is characterized by lack of chlorophyll and results in photosynthesis impairment, abnormal plant development and premature death. These abnormalities are frequently encountered in interspecific crosses and tissue culture experiments. Analysis of albino mutant phenotypes with full or partial chlorophyll deficiency can shed light on genetic determinants and molecular mechanisms of albinism. Here we report analysis of RNA-seq transcription profiling of barley (Hordeum vulgare L.) near-isogenic lines, one of which is a carrier of mutant allele of the Alm gene for albino lemma and pericarp phenotype (line i:BwAlm).
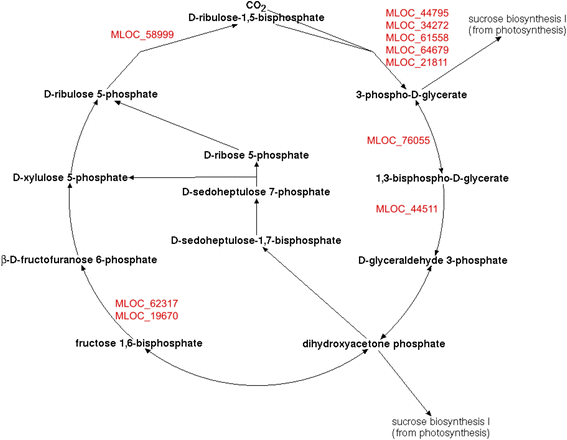
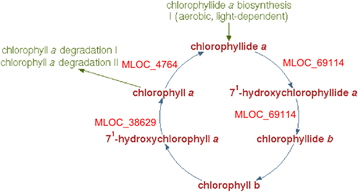
Results: 1221 genome fragments have statistically significant changes in expression levels between lines i:BwAlm and Bowman, with 148 fragments having increased expression levels in line i:BwAlm, and 1073 genome fragments, including 42 plastid operons, having decreased levels of expression in line i:BwAlm. We detected functional dissimilarity between genes with higher and lower levels of expression in i:BwAlm line. Genes with lower level of expression in the i:BwAlm line are mostly associated with photosynthesis and chlorophyll synthesis, while genes with higher expression level are functionally associated with vesicle transport. Differentially expressed genes are shown to be involved in several metabolic pathways; the largest fraction of such genes was observed for the Calvin-Benson-Bassham cycle. Finally, de novo assembly of transcriptome contains several transcripts, not annotated in current H. vulgare genome version.
Conclusions: Our results provide the new information about genes which could be involved in formation of albino lemma and pericarp phenotype. They demonstrate the interplay between nuclear and chloroplast genomes in this physiological process.
Keywords: Albino lemma; Barley; Chlorophyll synthesis; Differential expression; Gene network; IonTorrent sequencing platform; Near-isogenic lines; Nuclear genes; RNA-seq.
Figures











References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

