A conserved TLR5 binding and activation hot spot on flagellin
- PMID: 28106112
- PMCID: PMC5247705
- DOI: 10.1038/srep40878
A conserved TLR5 binding and activation hot spot on flagellin
Abstract
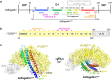
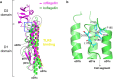
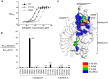
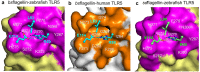
Flagellin is a bacterial protein that polymerizes into the flagellar filament and is essential for bacterial motility. When flagellated bacteria invade the host, flagellin is recognized by Toll-like receptor 5 (TLR5) as a pathogen invasion signal and eventually evokes the innate immune response. Here, we provide a conserved structural mechanism by which flagellins from Gram-negative γ-proteobacteria and Gram-positive Firmicutes bacteria bind and activate TLR5. The comparative structural analysis using our crystal structure of a complex between Bacillus subtilis flagellin (bsflagellin) and TLR5 at 2.1 Å resolution, combined with the alanine scanning analysis of the binding interface, reveals a common hot spot in flagellin for TLR5 activation. An arginine residue (bsflagellin R89) of the flagellin D1 domain and its adjacent residues (bsflagellin E114 and L93) constitute a hot spot that provides shape and chemical complementarity to a cavity generated by the loop of leucine-rich repeat 9 in TLR5. In addition to the flagellin D1 domain, the D0 domain also contributes to TLR5 activity through structurally dispersed regions, but not a single focal area. These results establish the groundwork for the future design of flagellin-based therapeutics.
Figures


References
-
- Yonekura K., Maki-Yonekura S. & Namba K. Growth mechanism of the bacterial flagellar filament. Research in microbiology 153, 191–197 (2002). - PubMed
-
- Macnab R. M. Genetics and biogenesis of bacterial flagella. Annual review of genetics 26, 131–158 (1992). - PubMed
-
- Ramos H. C., Rumbo M. & Sirard J. C. Bacterial flagellins: mediators of pathogenicity and host immune responses in mucosa. Trends in microbiology 12, 509–517 (2004). - PubMed
-
- Takeda K., Kaisho T. & Akira S. Toll-like receptors. Annu Rev Immunol 21, 335–376 (2003). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

