Parenclitic Network Analysis of Methylation Data for Cancer Identification
- PMID: 28107365
- PMCID: PMC5249089
- DOI: 10.1371/journal.pone.0169661
Parenclitic Network Analysis of Methylation Data for Cancer Identification
Abstract
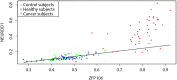
We make use of ideas from the theory of complex networks to implement a machine learning classification of human DNA methylation data, that carry signatures of cancer development. The data were obtained from patients with various kinds of cancers and represented as parenclictic networks, wherein nodes correspond to genes, and edges are weighted according to pairwise variation from control group subjects. We demonstrate that for the 10 types of cancer under study, it is possible to obtain a high performance of binary classification between cancer-positive and negative samples based on network measures. Remarkably, an accuracy as high as 93-99% is achieved with only 12 network topology indices, in a dramatic reduction of complexity from the original 15295 gene methylation levels. Moreover, it was found that the parenclictic networks are scale-free in cancer-negative subjects, and deviate from the power-law node degree distribution in cancer. The node centrality ranking and arising modular structure could provide insights into the systems biology of cancer.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

