VAPs and ACBD5 tether peroxisomes to the ER for peroxisome maintenance and lipid homeostasis
- PMID: 28108526
- PMCID: PMC5294787
- DOI: 10.1083/jcb.201608128
VAPs and ACBD5 tether peroxisomes to the ER for peroxisome maintenance and lipid homeostasis
Abstract
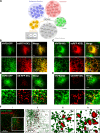
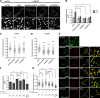
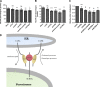
Lipid exchange between the endoplasmic reticulum (ER) and peroxisomes is necessary for the synthesis and catabolism of lipids, the trafficking of cholesterol, and peroxisome biogenesis in mammalian cells. However, how lipids are exchanged between these two organelles is not understood. In this study, we report that the ER-resident VAMP-associated proteins A and B (VAPA and VAPB) interact with the peroxisomal membrane protein acyl-CoA binding domain containing 5 (ACBD5) and that this interaction is required to tether the two organelles together, thereby facilitating the lipid exchange between them. Depletion of either ACBD5 or VAP expression results in increased peroxisome mobility, suggesting that VAP-ACBD5 complex acts as the primary ER-peroxisome tether. We also demonstrate that tethering of peroxisomes to the ER is necessary for peroxisome growth, the synthesis of plasmalogen phospholipids, and the maintenance of cellular cholesterol levels. Collectively, our data highlight the importance of VAP-ACBD5-mediated contact between the ER and peroxisomes for organelle maintenance and lipid homeostasis.
© 2017 Hua et al.
Figures


Comment in
-
Incredibly close-A newly identified peroxisome-ER contact site in humans.J Cell Biol. 2017 Feb;216(2):287-289. doi: 10.1083/jcb.201701072. Epub 2017 Jan 20. J Cell Biol. 2017. PMID: 28108527 Free PMC article.
-
Organelle dynamics: Connections, connections, connections.Nat Rev Mol Cell Biol. 2017 Feb 21;18(3):139. doi: 10.1038/nrm.2017.14. Nat Rev Mol Cell Biol. 2017. PMID: 28220048 No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

