Confirmation of five novel susceptibility loci for systemic lupus erythematosus (SLE) and integrated network analysis of 82 SLE susceptibility loci
- PMID: 28108556
- PMCID: PMC5731438
- DOI: 10.1093/hmg/ddx026
Confirmation of five novel susceptibility loci for systemic lupus erythematosus (SLE) and integrated network analysis of 82 SLE susceptibility loci
Abstract
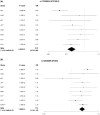
We recently identified ten novel SLE susceptibility loci in Asians and uncovered several additional suggestive loci requiring further validation. This study aimed to replicate five of these suggestive loci in a Han Chinese cohort from Hong Kong, followed by meta-analysis (11,656 cases and 23,968 controls) on previously reported Asian and European populations, and to perform bioinformatic analyses on all 82 reported SLE loci to identify shared regulatory signatures. We performed a battery of analyses for these five loci, as well as joint analyses on all 82 SLE loci. All five loci passed genome-wide significance: MYNN (rs10936599, Pmeta = 1.92 × 10-13, OR = 1.14), ATG16L2 (rs11235604, Pmeta = 8.87 × 10 -12, OR = 0.78), CCL22 (rs223881, Pmeta = 5.87 × 10-16, OR = 0.87), ANKS1A (rs2762340, Pmeta = 4.93 × 10-15, OR = 0.87) and RNASEH2C (rs1308020, Pmeta = 2.96 × 10-19, OR = 0.84) and co-located with annotated gene regulatory elements. The novel loci share genetic signatures with other reported SLE loci, including effects on gene expression, transcription factor binding, and epigenetic characteristics. Most (56%) of the correlated (r2 > 0.8) SNPs from the 82 SLE loci were implicated in differential expression (9.81 × 10-198 < P < 5 × 10-3) of cis-genes. Transcription factor binding sites for p53, MEF2A and E2F1 were significantly (P < 0.05) over-represented in SLE loci, consistent with apoptosis playing a critical role in SLE. Enrichment analysis revealed common pathways, gene ontology, protein domains, and cell type-specific expression. In summary, we provide evidence of five novel SLE susceptibility loci. Integrated bioinformatics using all 82 loci revealed that SLE susceptibility loci share many gene regulatory features, suggestive of conserved mechanisms of SLE etiopathogenesis.
© The Author 2017. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures



References
-
- Alarcon G.S., McGwin G. Jr., Petri M., Reveille J.D., Ramsey-Goldman R., Kimberly R.P. and PROFILE Study Group (2002) Baseline characteristics of a multiethnic lupus cohort: PROFILE. Lupus, 11, 95–101. - PubMed
-
- International Consortium for Systemic Lupus Erythematosus, G., Harley J.B., Alarcon-Riquelme M.E., Criswell L.A., Jacob C.O., Kimberly R.P., Moser K.L., Tsao B.P., Vyse T.J., Langefeld C.D., et al. (2008) Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat. Genet., 40, 204–210. - PMC - PubMed
-
- Bentham J., Morris D.L., Cunninghame Graham D.S., Pinder C.L., Tombleson P., Behrens T.W., Martin J., Fairfax B.P., Knight J.C., Chen L., et al. (2015) Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat. Genet., 47, 1457–1464. - PMC - PubMed
-
- Morris D.L., Sheng Y., Zhang Y., Wang Y.F., Zhu Z., Tombleson P., Chen L., Cunninghame Graham D.S., Bentham J., Roberts A.L., et al. (2016) Genome-wide association meta-analysis in Chinese and European individuals identifies ten new loci associated with systemic lupus erythematosus. Nat. Genet., 48,940–946. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

