Revealing the role of the product metal in DNA polymerase β catalysis
- PMID: 28108654
- PMCID: PMC5389463
- DOI: 10.1093/nar/gkw1363
Revealing the role of the product metal in DNA polymerase β catalysis
Abstract
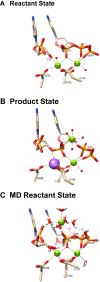
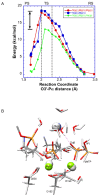
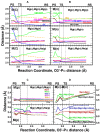
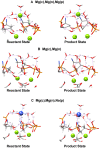
DNA polymerases catalyze a metal-dependent nucleotidyl transferase reaction during extension of a DNA strand using the complementary strand as a template. The reaction has long been considered to require two magnesium ions. Recently, a third active site magnesium ion was identified in some DNA polymerase product crystallographic structures, but its role is not known. Using quantum mechanical/ molecular mechanical calculations of polymerase β, we find that a third magnesium ion positioned near the newly identified product metal site does not alter the activation barrier for the chemical reaction indicating that it does not have a role in the forward reaction. This is consistent with time-lapse crystallographic structures following insertion of Sp-dCTPαS. Although sulfur substitution deters product metal binding, this has only a minimal effect on the rate of the forward reaction. Surprisingly, monovalent sodium or ammonium ions, positioned in the product metal site, lowered the activation barrier. These calculations highlight the impact that an active site water network can have on the energetics of the forward reaction and how metals or enzyme side chains may interact with the network to modulate the reaction barrier. These results also are discussed in the context of earlier findings indicating that magnesium at the product metal position blocks the reverse pyrophosphorolysis reaction.
Published by Oxford University Press on behalf of Nucleic Acids Research 2017.
Figures


Similar articles
-
Requirement for transient metal ions revealed through computational analysis for DNA polymerase going in reverse.Proc Natl Acad Sci U S A. 2015 Sep 22;112(38):E5228-36. doi: 10.1073/pnas.1511207112. Epub 2015 Sep 8. Proc Natl Acad Sci U S A. 2015. PMID: 26351676 Free PMC article.
-
Critical role of magnesium ions in DNA polymerase beta's closing and active site assembly.J Am Chem Soc. 2004 Jul 14;126(27):8441-53. doi: 10.1021/ja049412o. J Am Chem Soc. 2004. PMID: 15238001
-
Crystallographic evidence for two-metal-ion catalysis in human pol η.Protein Sci. 2019 Feb;28(2):439-447. doi: 10.1002/pro.3541. Epub 2018 Dec 11. Protein Sci. 2019. PMID: 30368948 Free PMC article.
-
New structural snapshots provide molecular insights into the mechanism of high fidelity DNA synthesis.DNA Repair (Amst). 2015 Aug;32:3-9. doi: 10.1016/j.dnarep.2015.04.007. Epub 2015 Apr 30. DNA Repair (Amst). 2015. PMID: 26002198 Free PMC article. Review.
-
Kinetic Mechanism of DNA Polymerases: Contributions of Conformational Dynamics and a Third Divalent Metal Ion.Chem Rev. 2018 Jun 27;118(12):6000-6025. doi: 10.1021/acs.chemrev.7b00685. Epub 2018 Jun 4. Chem Rev. 2018. PMID: 29863852 Review.
Cited by
-
A non-natural nucleotide uses a specific pocket to selectively inhibit telomerase activity.PLoS Biol. 2019 Apr 5;17(4):e3000204. doi: 10.1371/journal.pbio.3000204. eCollection 2019 Apr. PLoS Biol. 2019. PMID: 30951520 Free PMC article.
-
Combining Evolutionary Conservation and Quantum Topological Analyses To Determine Quantum Mechanics Subsystems for Biomolecular Quantum Mechanics/Molecular Mechanics Simulations.J Chem Theory Comput. 2021 Jul 13;17(7):4524-4537. doi: 10.1021/acs.jctc.1c00313. Epub 2021 Jun 4. J Chem Theory Comput. 2021. PMID: 34087064 Free PMC article.
-
Catalytic mechanism of DNA polymerases-Two metal ions or three?Protein Sci. 2019 Feb;28(2):288-291. doi: 10.1002/pro.3542. Epub 2018 Dec 20. Protein Sci. 2019. PMID: 30368961 Free PMC article. Review. No abstract available.
-
Time-lapse crystallography snapshots of a double-strand break repair polymerase in action.Nat Commun. 2017 Aug 15;8(1):253. doi: 10.1038/s41467-017-00271-7. Nat Commun. 2017. PMID: 28811466 Free PMC article.
-
Watching a double strand break repair polymerase insert a pro-mutagenic oxidized nucleotide.Nat Commun. 2021 Apr 6;12(1):2059. doi: 10.1038/s41467-021-21354-6. Nat Commun. 2021. PMID: 33824325 Free PMC article.
References
-
- Bebenek K., Kunkel T.A.. Functions of DNA polymerases. Adv Protein Chem. 2004; 69:137–165. - PubMed
-
- Beard W.A., Shock D.D., Vande Berg B.J., Wilson S.H.. Efficiency of correct nucleotide insertion governs DNA polymerase fidelity. J. Biol. Chem. 2002; 277:47393–47398. - PubMed
-
- Pelletier H., Sawaya M.R., Kumar A., Wilson S.H., Kraut J.. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994; 264:1891–1903. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

