Tumor evolution: Linear, branching, neutral or punctuated?
- PMID: 28110020
- PMCID: PMC5558210
- DOI: 10.1016/j.bbcan.2017.01.003
Tumor evolution: Linear, branching, neutral or punctuated?
Abstract
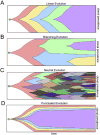
Intratumor heterogeneity has been widely reported in human cancers, but our knowledge of how this genetic diversity emerges over time remains limited. A central challenge in studying tumor evolution is the difficulty in collecting longitudinal samples from cancer patients. Consequently, most studies have inferred tumor evolution from single time-point samples, providing very indirect information. These data have led to several competing models of tumor evolution: linear, branching, neutral and punctuated. Each model makes different assumptions regarding the timing of mutations and selection of clones, and therefore has different implications for the diagnosis and therapeutic treatment of cancer patients. Furthermore, emerging evidence suggests that models may change during tumor progression or operate concurrently for different classes of mutations. Finally, we discuss data that supports the theory that most human tumors evolve from a single cell in the normal tissue. This article is part of a Special Issue entitled: Evolutionary principles - heterogeneity in cancer?, edited by Dr. Robert A. Gatenby.
Keywords: Cancer biology; Cancer genomics; Genome evolution; Intratumor heterogeneity; Single cell genomics; Tumor evolution.
Copyright © 2017 Elsevier B.V. All rights reserved.
Figures



References
-
- Bignold LP, Coghlan BLD, Jersmann HPA. Hansemann’s Ideas of the Nature of Cancer: Description and Analysis, David Paul Von Hansemann: Contributions to Oncology, Birkhäuser. Basel. 2007:75–90.
-
- Di Vinci A, Infusini E, Peveri C, Sciutto A, Orecchia R, Geido E, Monaco R, Giaretti W. Intratumor heterogeneity of chromosome 1, 7, 17, and 18 aneusomies obtained by FISH and association with flow cytometric DNA index in human colorectal adenocarcinomas. Cytometry. 1999;35:369–375. - PubMed
-
- Szollosi J, Balazs M, Feuerstein BG, Benz CC, Waldman FM. ERBB-2 (HER2/neu) gene copy number, p185HER-2 overexpression, and intratumor heterogeneity in human breast cancer. Cancer Res. 1995;55:5400–5407. - PubMed
-
- Farabegoli F, Santini D, Ceccarelli C, Taffurelli M, Marrano D, Baldini N. Clone heterogeneity in diploid and aneuploid breast carcinomas as detected by FISH. Cytometry. 2001;46:50–56. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

