Mitonuclear Interactions Mediate Transcriptional Responses to Hypoxia in Drosophila
- PMID: 28110272
- PMCID: PMC6095086
- DOI: 10.1093/molbev/msw246
Mitonuclear Interactions Mediate Transcriptional Responses to Hypoxia in Drosophila
Abstract
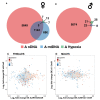
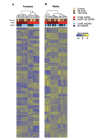
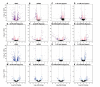
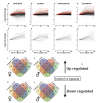
Among the major challenges in quantitative genetics and personalized medicine is to understand how gene × gene interactions (G × G: epistasis) and gene × environment interactions (G × E) underlie phenotypic variation. Here, we use the intimate relationship between mitochondria and oxygen availability to dissect the roles of nuclear DNA (nDNA) variation, mitochondrial DNA (mtDNA) variation, hypoxia, and their interactions on gene expression in Drosophila melanogaster. Mitochondria provide an important evolutionary and medical context for understanding G × G and G × E given their central role in integrating cellular signals. We hypothesized that hypoxia would alter mitonuclear communication and gene expression patterns. We show that first order nDNA, mtDNA, and hypoxia effects vary between the sexes, along with mitonuclear epistasis and G × G × E effects. Females were generally more sensitive to genetic and environmental perturbation. While dozens to hundreds of genes are altered by hypoxia in individual genotypes, we found very little overlap among mitonuclear genotypes for genes that were significantly differentially expressed as a consequence of hypoxia; excluding the gene hairy. Oxidative phosphorylation genes were among the most influenced by hypoxia and mtDNA, and exposure to hypoxia increased the signature of mtDNA effects, suggesting retrograde signaling between mtDNA and nDNA. We identified nDNA-encoded genes in the electron transport chain (succinate dehydrogenase) that exhibit female-specific mtDNA effects. Our findings have important implications for personalized medicine, the sex-specific nature of mitonuclear communication, and gene × gene coevolution under variable or changing environments.
Keywords: Drosophila; E; G ×; epistasis; hypoxia; mitonuclear; mtDNA; transcriptome.
© The Author 2016. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
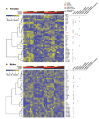

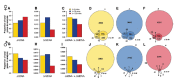

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

