Human iPSC-Derived Cerebral Organoids Model Cellular Features of Lissencephaly and Reveal Prolonged Mitosis of Outer Radial Glia
- PMID: 28111201
- PMCID: PMC5667944
- DOI: 10.1016/j.stem.2016.12.007
Human iPSC-Derived Cerebral Organoids Model Cellular Features of Lissencephaly and Reveal Prolonged Mitosis of Outer Radial Glia
Abstract
Classical lissencephaly is a genetic neurological disorder associated with mental retardation and intractable epilepsy, and Miller-Dieker syndrome (MDS) is the most severe form of the disease. In this study, to investigate the effects of MDS on human progenitor subtypes that control neuronal output and influence brain topology, we analyzed cerebral organoids derived from control and MDS-induced pluripotent stem cells (iPSCs) using time-lapse imaging, immunostaining, and single-cell RNA sequencing. We saw a cell migration defect that was rescued when we corrected the MDS causative chromosomal deletion and severe apoptosis of the founder neuroepithelial stem cells, accompanied by increased horizontal cell divisions. We also identified a mitotic defect in outer radial glia, a progenitor subtype that is largely absent from lissencephalic rodents but critical for human neocortical expansion. Our study, therefore, deepens our understanding of MDS cellular pathogenesis and highlights the broad utility of cerebral organoids for modeling human neurodevelopmental disorders.
Keywords: cerebral organoids; human lissencephaly; migration; outer radial glia; spindle orientation.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures

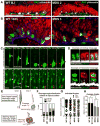

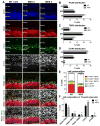

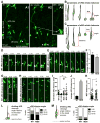
Comment in
-
Of Mice and Men: Species-Specific Organoid Models of Neocortical Malformation.Cell Stem Cell. 2017 Apr 6;20(4):421-422. doi: 10.1016/j.stem.2017.03.005. Cell Stem Cell. 2017. PMID: 28388422
-
You Have Brains in Your Head, You Have Organoids in Your Dish, You Can Steer Yourself in any Direction You Wish.Epilepsy Curr. 2017 Sep-Oct;17(5):311-313. doi: 10.5698/1535-7597.17.5.311. Epilepsy Curr. 2017. PMID: 29225549 Free PMC article. No abstract available.
References
-
- Armstrong E, Schleicher A, Omran H, Curtis M, Zilles K. The ontogeny of human gyrification. Cereb Cortex. 1995;1:56–63. - PubMed
-
- Barkovich AJ, Koch TK, Carrol CL. The spectrum of lissencephaly: report of ten cases analyzed by magnetic resonance imaging. Ann Neurol. 1991;30:139–146. - PubMed
-
- Betizeau M, Cortay V, Patti D, Pfister S, Gautier E, Bellemin-Ménard A, Afanassieff M, Huissoud C, Douglas RJ, Kennedy H, Dehay C. Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron. 2013;2:442–57. - PubMed
-
- Borrell V, Gotz M. Role of radial glial cells in cerebral cortex folding. Curr Opin Neurobiol. 2014;27:39–46. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

