Genome and transcriptome analysis of surfactin biosynthesis in Bacillus amyloliquefaciens MT45
- PMID: 28112210
- PMCID: PMC5256033
- DOI: 10.1038/srep40976
Genome and transcriptome analysis of surfactin biosynthesis in Bacillus amyloliquefaciens MT45
Abstract
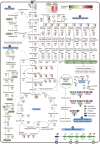
Natural Bacillus isolates generate limited amounts of surfactin (<10% of their biomass), which functions as an antibiotic or signalling molecule in inter-/intra-specific interactions. However, overproduction of surfactin in Bacillus amyloliquefaciens MT45 was observed at a titre of 2.93 g/l, which is equivalent to half of the maximum biomass. To systemically unravel this efficient biosynthetic process, the genome and transcriptome of this bacterium were compared with those of B. amyloliquefaciens type strain DSM7T. MT45 possesses a smaller genome while containing more unique transporters and resistance-associated genes. Comparative transcriptome analysis revealed notable enrichment of the surfactin synthesis pathway in MT45, including central carbon metabolism and fatty acid biosynthesis to provide sufficient quantities of building precursors. Most importantly, the modular surfactin synthase overexpressed (9 to 49-fold) in MT45 compared to DSM7T suggested efficient surfactin assembly and resulted in the overproduction of surfactin. Furthermore, based on the expression trends observed in the transcriptome, there are multiple potential regulatory genes mediating the expression of surfactin synthase. Thus, the results of the present study provide new insights regarding the synthesis and regulation of surfactin in high-producing strain and enrich the genomic and transcriptomic resources available for B. amyloliquefaciens.
Figures




References
-
- Ogura M., Yamaguchi H., Yoshida K.-i., Fujita Y. & Tanaka T. DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of B. subtilis two-component regulatory systems. Nucleic Acids Res. 29, 3804–3813, doi: 10.1093/nar/29.18.3804 (2001). - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

