Multi-omics analyses reveal metabolic alterations regulated by hepatitis B virus core protein in hepatocellular carcinoma cells
- PMID: 28112229
- PMCID: PMC5253728
- DOI: 10.1038/srep41089
Multi-omics analyses reveal metabolic alterations regulated by hepatitis B virus core protein in hepatocellular carcinoma cells
Abstract
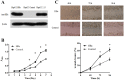
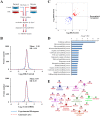
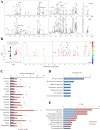
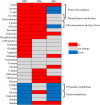
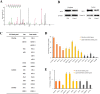
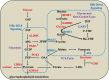
Chronic hepatitis B virus (HBV) infection is partly responsible for hepatitis, fatty liver disease and hepatocellular carcinoma (HCC). HBV core protein (HBc), encoded by the HBV genome, may play a significant role in HBV life cycle. However, the function of HBc in the occurrence and development of liver disease is still unclear. To investigate the underlying mechanisms, HBc-transfected HCC cells were characterized by multi-omics analyses. Combining proteomics and metabolomics analyses, our results showed that HBc promoted the expression of metabolic enzymes and the secretion of metabolites in HCC cells. In addition, glycolysis and amino acid metabolism were significantly up-regulated by HBc. Moreover, Max-like protein X (MLX) might be recruited and enriched by HBc in the nucleus to regulate glycolysis pathways. This study provides further insights into the function of HBc in the molecular pathogenesis of HBV-induced diseases and indicates that metabolic reprogramming appears to be a hallmark of HBc transfection.
Figures

References
-
- Hanahan D. & Weinberg R. A. The hallmarks of cancer. Cell 100, 57–70 (2000). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

