SecretEPDB: a comprehensive web-based resource for secreted effector proteins of the bacterial types III, IV and VI secretion systems
- PMID: 28112271
- PMCID: PMC5253721
- DOI: 10.1038/srep41031
SecretEPDB: a comprehensive web-based resource for secreted effector proteins of the bacterial types III, IV and VI secretion systems
Abstract
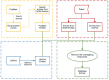
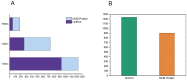
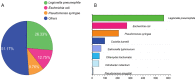
Bacteria translocate effector molecules to host cells through highly evolved secretion systems. By definition, the function of these effector proteins is to manipulate host cell biology and the sequence, structural and functional annotations of these effector proteins will provide a better understanding of how bacterial secretion systems promote bacterial survival and virulence. Here we developed a knowledgebase, termed SecretEPDB (Bacterial Secreted Effector Protein DataBase), for effector proteins of type III secretion system (T3SS), type IV secretion system (T4SS) and type VI secretion system (T6SS). SecretEPDB provides enriched annotations of the aforementioned three classes of effector proteins by manually extracting and integrating structural and functional information from currently available databases and the literature. The database is conservative and strictly curated to ensure that every effector protein entry is supported by experimental evidence that demonstrates it is secreted by a T3SS, T4SS or T6SS. The annotations of effector proteins documented in SecretEPDB are provided in terms of protein characteristics, protein function, protein secondary structure, Pfam domains, metabolic pathway and evolutionary details. It is our hope that this integrated knowledgebase will serve as a useful resource for biological investigation and the generation of new hypotheses for research efforts aimed at bacterial secretion systems.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

