Automatic Segmentation and Quantification of White and Brown Adipose Tissues from PET/CT Scans
- PMID: 28114010
- PMCID: PMC6421081
- DOI: 10.1109/TMI.2016.2636188
Automatic Segmentation and Quantification of White and Brown Adipose Tissues from PET/CT Scans
Abstract
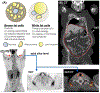
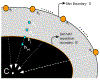
In this paper, we investigate the automatic detection of white and brown adipose tissues using Positron Emission Tomography/Computed Tomography (PET/CT) scans, and develop methods for the quantification of these tissues at the whole-body and body-region levels. We propose a patient-specific automatic adiposity analysis system with two modules. In the first module, we detect white adipose tissue (WAT) and its two sub-types from CT scans: Visceral Adipose Tissue (VAT) and Subcutaneous Adipose Tissue (SAT). This process relies conventionally on manual or semi-automated segmentation, leading to inefficient solutions. Our novel framework addresses this challenge by proposing an unsupervised learning method to separate VAT from SAT in the abdominal region for the clinical quantification of central obesity. This step is followed by a context driven label fusion algorithm through sparse 3D Conditional Random Fields (CRF) for volumetric adiposity analysis. In the second module, we automatically detect, segment, and quantify brown adipose tissue (BAT) using PET scans because unlike WAT, BAT is metabolically active. After identifying BAT regions using PET, we perform a co-segmentation procedure utilizing asymmetric complementary information from PET and CT. Finally, we present a new probabilistic distance metric for differentiating BAT from non-BAT regions. Both modules are integrated via an automatic body-region detection unit based on one-shot learning. Experimental evaluations conducted on 151 PET/CT scans achieve state-of-the-art performances in both central obesity as well as brown adiposity quantification.
Figures





References
-
- Preston SH, “Deadweight?—The influence of obesity on longevity,” N. Eng. J. Med, vol. 352, no. 11, pp. 1135–1137, 2005. - PubMed
-
- Cefalu WT et al. , “Insulin resistance and fat patterning with aging: Relationship to metabolic risk factors for cardiovascular disease,” Metabolism, vol. 47, no. 4, pp. 401–408, April 1998. - PubMed
-
- Gastaldelli A et al. , “Metabolic effects of visceral fat accumulation in type 2 diabetes,” J. Clin. Endocrinol. Metabolism, vol. 87, no. 11, pp. 5098–5103, 2002. - PubMed
-
- National Center for Health Statistics and Centers for Disease Control and Prevention: Health, United States, With Special Feature on Racial and Ethnic Health Disparities, 2015. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

