Scalable Parameter Estimation for Genome-Scale Biochemical Reaction Networks
- PMID: 28114351
- PMCID: PMC5256869
- DOI: 10.1371/journal.pcbi.1005331
Scalable Parameter Estimation for Genome-Scale Biochemical Reaction Networks
Abstract
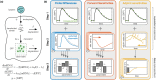
Mechanistic mathematical modeling of biochemical reaction networks using ordinary differential equation (ODE) models has improved our understanding of small- and medium-scale biological processes. While the same should in principle hold for large- and genome-scale processes, the computational methods for the analysis of ODE models which describe hundreds or thousands of biochemical species and reactions are missing so far. While individual simulations are feasible, the inference of the model parameters from experimental data is computationally too intensive. In this manuscript, we evaluate adjoint sensitivity analysis for parameter estimation in large scale biochemical reaction networks. We present the approach for time-discrete measurement and compare it to state-of-the-art methods used in systems and computational biology. Our comparison reveals a significantly improved computational efficiency and a superior scalability of adjoint sensitivity analysis. The computational complexity is effectively independent of the number of parameters, enabling the analysis of large- and genome-scale models. Our study of a comprehensive kinetic model of ErbB signaling shows that parameter estimation using adjoint sensitivity analysis requires a fraction of the computation time of established methods. The proposed method will facilitate mechanistic modeling of genome-scale cellular processes, as required in the age of omics.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

